EconML
 EconML copied to clipboard
EconML copied to clipboard
Different features for model_y and model_t
Is it possible to use different features as inputs into model_y and model_t when using LinearDML?
Hi.Nice to meet you. I thought the same thing before, but I think the methods in this package probably don't support covariate selection (X in model_y and model_t should be shared).
However, if you mask unnecessary covariates with sklearn's ColumnTransformer, build a pipeline from feature selection to model adaptation with sklearn's pipeline, and pass it to DML's arguments, model_y and model_t. I think you can do practically the same thing.
for example:
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
preprocessor = ColumnTransformer(
transformers=[
("hoge1", hogehoge),
("hoge2", Pass),
]
)
model_t = Pipeline(
steps=[("preprocessor", preprocessor), ("classifier", LogisticRegression())]
)
https://scikit-learn.org/stable/auto_examples/compose/plot_column_transformer_mixed_types.html
@MasaAsami is correct that you can gain some control via preprocessing in a pipeline, but be aware that the features always include the columns of both X and W, and that if linear_first_stages is true then the input to the Y model also has interactions of those columns with the featurized version of X, so you would need to use some care when setting up the preprocessor.
Is there a particular reason you'd like to use different features in your setting?
Thenk you @kbattocchi .
Is there a particular reason you'd like to use different features in your setting?
In practical terms, I don't think it is necessary to separate the covariates.
However, for my theoretical confirmation, I was thinking of swapping the covariates to check the behavior of the final estimates (I've been busy, so I haven't been able to check yet...)
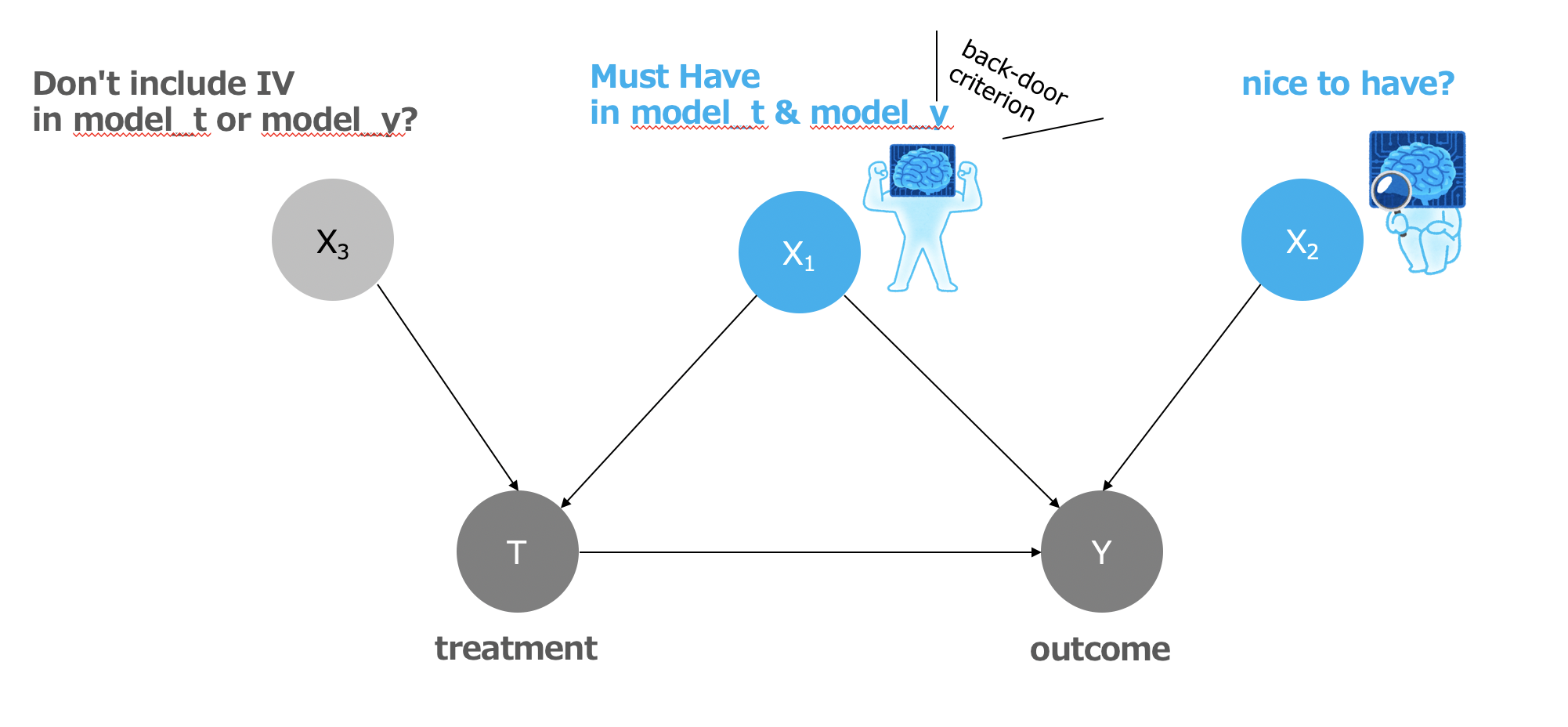
In particular, I was thinking of testing X2 as shown in the DAG below, with four patterns: model_y only, model_t only, both models included, and both models not included, to verify the final ATE estimates and variances. I think that the pattern that includes both models is probably the best, but I haven't checked it yet.