ProDy

ProDy copied to clipboard
added gromos rmsd clustering
This method is the same as the one used in https://manual.gromacs.org/documentation/current/onlinehelp/gmx-cluster.html
Perhaps we should give this a better name
Definitely, something isn't right here for the labels=None case. Perhaps make labels a necessary argument rather than defaulting to None
Actually, we do want to support the case where we have no labels, e.g. from an MD trajectory. It was just done wrong.
It should be good now, but needs testing
This works to cluster a set of trajectories now:
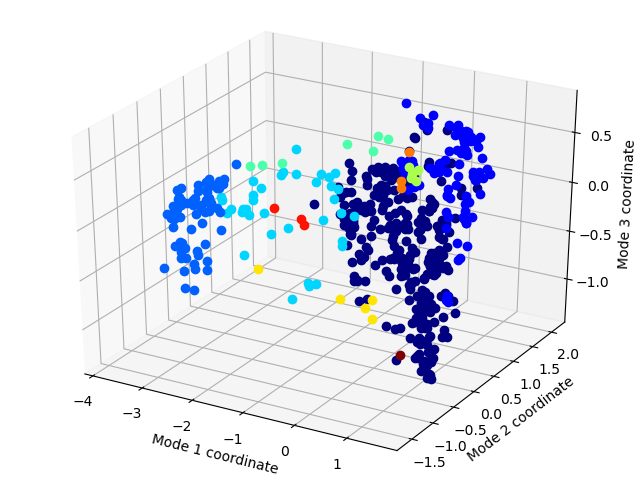
from prody import *
import matplotlib.pyplot as plt
plt.ion()
import numpy as np
ref_pca = loadModel('prody_pca/ref.pca.npz')
ens = parseDCD("combined.dcd", step=100)
ens.iterpose(rmsd=0.01)
rmsd_matrix = ens.getRMSDs(pairwise=True)
x = calcRMSDclusters(rmsd_matrix, 2.1)
colors = np.zeros(num_confs, dtype=int)
for i, c in enumerate(x):
colors[list(np.array(c, dtype=int))] = i
colors = list(colors)
plt.figure()
showProjection(ens, ref_pca[:3], markersize=3, color=colors)
plt.savefig('gromos_clusters_2p1A.png')