Insonsistent / incorrect handling of OME modulo structured annoations
I am looking at the modulo-Datasets here: https://docs.openmicroscopy.org/ome-model/6.0.0/ome-tiff/data.html#modulo-datasets
Which use the XML structured annotation documented here: https://docs.openmicroscopy.org/ome-model/6.0.0/developers/6d-7d-and-8d-storage.html
Opening the files with FIJI via the Bioformats plugins seems to show some support for the annotations but it seems to be mostly broken / inconsistent.
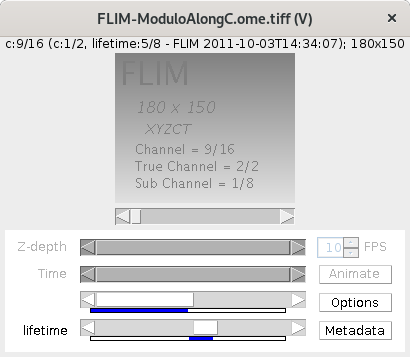 Looking at
Looking at FLIM-ModuloAlongC.ome.tiffwith the Hyperstack viewer; the lifetime index displayed above the image does not correspond with the Sub Channel displayed in the image itself. Using the Data Browser viewer a lifetime-Slider is displayed that corresponds with the lifetime-Index displayed above the image but does also not correspond with the Sub Channel displayed in the image (e.g. lifetime 5/8; Sub Channel 1/8).
 Looking at
Looking at FLIM-ModuloAlongT-TSCPC.ome.tiff there is no lifetime-Index above the image nor is there a lifetime-Slider in the Data Browser.
Thanks for the report @ngladitz, I can confirm that the display handler is not properly interpreting the Sub Channels, the text imprinted in the image is the correct value. Also at the moment the plugins display is only handling Modulo C, so support for Modulo T and Modulo Z will need to be added also.
I looked into this bug and I am quite sure the bug is in positionToRaster:
https://github.com/ome/bioformats/blob/b8b9d472ffe3c8c29972b2e44ae4a41d6f873e70/components/formats-api/src/loci/formats/FormatTools.java#L750
The loop should start with the modulo index instead of the true index:
public static int positionToRaster(int[] lengths, int[] pos) {
int offset = 1;
int raster = 0;
// for (int i=0; i<pos.length; i++) {
for (int i=pos.length - 1; i >= 0; i--) {
raster += offset * pos[i];
offset *= lengths[i];
}
return raster;
}
@alex-hildebrand, if you have a set of changes that fix this issue, you're welcome to open a pull request for review. If you choose to open a pull request, please note that we require a Contributor License Agreement in order to accept any changes.
Note though that positionToRaster does not (and is not intended) to have any awareness of specific dimensions, including modulo dimensions. The length and order of any dimensions are encoded in the lengths parameter. Typically when there is a problem with how positionToRaster behaves, it has to do with incorrect arrays being passed in.