Why is some vertices zeros in den-91k space?
Hello,
I have completed the resting-state fMRI preprocessing based on fmriprep 20.1.1. When I checked the results in den-91k space (i.e., files named ...fsLR_den-91k_bold.dtseries.nii), I found there are 618 vertices being zero in these files. Specifically, 295 vertices on the left surface, 323 vertices on the right surface, and no subcortical volumetric voxels. For each subject, it appear that the indices of these zero vertices is same. But I also checked the files in HCP database, all value (91282 vertices/voxels) are nonzero. So why? How do we deal with these vertices? And are there any commands (e.g., any functions of MySQL Workbench) to fit/interpolate these vertices?
Besides, what's the resampling/registering process between different output space? For example, is fsaverage6 space file registerd from original-space BOLD file or from standard-space BOLD file such MNI152NLin2009cAsym ? Is fsaverage5 file resampled from fsaverage6 or fsaverage file? Is den-91k space file resampled from fsaverage file or registed from original-space BOLD file?
Finally, which BOLD space do WM signals and CSF siginals (in ...confounds_regressors.tsv) extract from?
Thanks a lot!
Hi @ly6ustc ,
Are you processing the same data you are comparing within the HCP database?
This may be related to https://github.com/nipreps/fmriprep/issues/2333 - two things to that come to mind: is your data limited FOV or does the fMRIPrep brain mask have some dropout?
Surface BOLD time-series are generated from the BOLD image in T1w space - when producing CIFTI, an extra resampling step is added. The surface CIFTI generation goes:
BOLD in T1w -> fsaverage (via mri_vol2surf) -> fsLR (via wb_command -metric-resample)
You may want to verify the 0-value vertices are present in the fsaverage GIfTIs produced, to ensure this wasn't introduced on the fsLR resampling step.
Confounds are extracted from the native BOLD space.
Hi @ly6ustc ,
Are you processing the same data you are comparing within the HCP database?
This may be related to #2333 - two things to that come to mind: is your data limited FOV or does the fMRIPrep brain mask have some dropout?
Surface BOLD time-series are generated from the BOLD image in T1w space - when producing CIFTI, an extra resampling step is added. The surface CIFTI generation goes:
BOLD in T1w -> fsaverage (via mri_vol2surf) -> fsLR (via wb_command -metric-resample)You may want to verify the 0-value vertices are present in the fsaverage GIfTIs produced, to ensure this wasn't introduced on the fsLR resampling step.
Confounds are extracted from the native BOLD space.
Thanks for your responses. First, I didn't use fmriprep on HCP data. So maybe it's unfair to compare them. Actually, I just wanted to make sure if all vertex of 91k space are nonzero.
I have checked some subjects on my dataset. Their FOV is Ok, containing the whole brain. I don't unsderstand your seconde hypothesis,"does the fMRIPrep brain mask have some dropout?" So I displayed two pictures here, representing "sub-xxx_task-rest_space-T1w_boldref.nii.gz" and "sub-xxx_task-rest_space-T1w_desc-brain_mask.nii.gz" in directory "fmriprep/sub-xxx/func/". I think they are fine.
I also checked the BOLD files in the fsaverage space. For fsaverage6, there are 3486 zero vertices in left cortex, and 3491 zero vertices in right cortex. These number is equal to the number of medial wall vertices. So did fsaverage5 space (888 vertices in left cortex and 881 vertices in right cortex). So I think the results of fsaverage is OK. I didn't observe more high resolution space because I didn't have these.
Furthermore, I have checked all results named sub-xxx_task-rest_space-fsLR_den-91k_bold.dtseries.nii of my dataset (from three different sites). They all contain 618 vertices of zero with the same indices. So I think the problems may be relate to resampling, is it possiple that fsLR resample more medial wall vertices from fsaverage? Or is it up to my fmriprep version (20.1.1)? Is it Ok (59,412 nonzero vertices in cortex) for others using fmriprep?
My fmriprep docker commands is :
docker run -i --name fmriprep$name --rm
-v $fs_licence:/opt/freesurfer/license.txt:ro
-v $BIDSdir:/data:ro
-v /home/z/Data/Toolbox/templateflow:/home/fmriprep/.cache/templateflow
-v $prepre_dir:/out
-v $work_dir/work:/scratch
poldracklab/fmriprep:latest
/data /out
participant --participant-label $name
-w /scratch
--use-aroma
--nthreads $nthreads
--output-spaces $out_space
--cifti-output
Thank you very much!
I also observed the position of these 618 vertices of zero. As shown in figure, they (red vertices in figure) are adjacent to medial wall (yellow vertices in figure). So I think fsLR resample more medial wall vertices from fsaverage.
Thanks for following up - I'm unsure if this is a bug or a limitation. fMRIPrep's version of CIFTIs have a few notable differences from ones generated by HCP Pipelines:
- No multimodal surface matching (MSM)
- No gaussian surface smoothing
Instead of MSM, we resample the FreeSurfer surfaces via their recommendations (PDF, Part A).
It seems others have run into a similar problem with zero values near the medial wall, and used wb_command -cifti-dilate to remedy it.
https://wiki.humanconnectome.org/display/PublicData/DIY+fix+for+zeroes+near+medial+wall+in+rfMRI+dtseries+data
Thanks for following up - I'm unsure if this is a bug or a limitation. fMRIPrep's version of CIFTIs have a few notable differences from ones generated by HCP Pipelines:
- No multimodal surface matching (MSM)
- No gaussian surface smoothing
Instead of MSM, we resample the FreeSurfer surfaces via their recommendations (PDF, Part A).
It seems others have run into a similar problem with zero values near the medial wall, and used
wb_command -cifti-dilateto remedy it. https://wiki.humanconnectome.org/display/PublicData/DIY+fix+for+zeroes+near+medial+wall+in+rfMRI+dtseries+data
Thank you very much, it works.
Hi @ly6ustc ,
Are you processing the same data you are comparing within the HCP database?
This may be related to #2333 - two things to that come to mind: is your data limited FOV or does the fMRIPrep brain mask have some dropout?
Surface BOLD time-series are generated from the BOLD image in T1w space - when producing CIFTI, an extra resampling step is added. The surface CIFTI generation goes:
BOLD in T1w -> fsaverage (via mri_vol2surf) -> fsLR (via wb_command -metric-resample)You may want to verify the 0-value vertices are present in the fsaverage GIfTIs produced, to ensure this wasn't introduced on the fsLR resampling step.
Confounds are extracted from the native BOLD space.
Hi @ly6ustc ,
Are you processing the same data you are comparing within the HCP database?
This may be related to #2333 - two things to that come to mind: is your data limited FOV or does the fMRIPrep brain mask have some dropout?
Surface BOLD time-series are generated from the BOLD image in T1w space - when producing CIFTI, an extra resampling step is added. The surface CIFTI generation goes:
BOLD in T1w -> fsaverage (via mri_vol2surf) -> fsLR (via wb_command -metric-resample)You may want to verify the 0-value vertices are present in the fsaverage GIfTIs produced, to ensure this wasn't introduced on the fsLR resampling step.
Confounds are extracted from the native BOLD space.
Hi @mgxd ,
I'm trying to extract some anatomical files such as thickness from Freesufer space to fsLR32K space. And I want to make this process consistent with what fmriprep's BOLD image did, i.e., “–cifti-output”. Based on your introduction, I used these codes below.
Step 1, resample the anatomical files of native space to group fsaverage space:
mri_surf2surf --srcsubject sub-F001 --trgsubject fsaverage --hemi lh --sval $path/sub-F001/surf/lh.thickness --tval $outpath/space-fsaverage_hemi-L.thickness.func.gii --cortex
Step2, resample the anatomical files of fsaverage space to fsLR32K space:
wb_command -metric-resample space-fsaverage_hemi-L.thickness.func.gii $current_sphere_L $target_sphere_L ADAP_BARY_AREA space-fsLR32K_hemi-L.thickness.func.gii -area-metrics $current_area_L $target_area_L
For $current_sphere_L, $target_sphere_L, $current_area_L, and $target_area_L, I used these templates in the yellow lines in the screenshot below.
Are right for these codes? Should I adjust some parameters in commands?
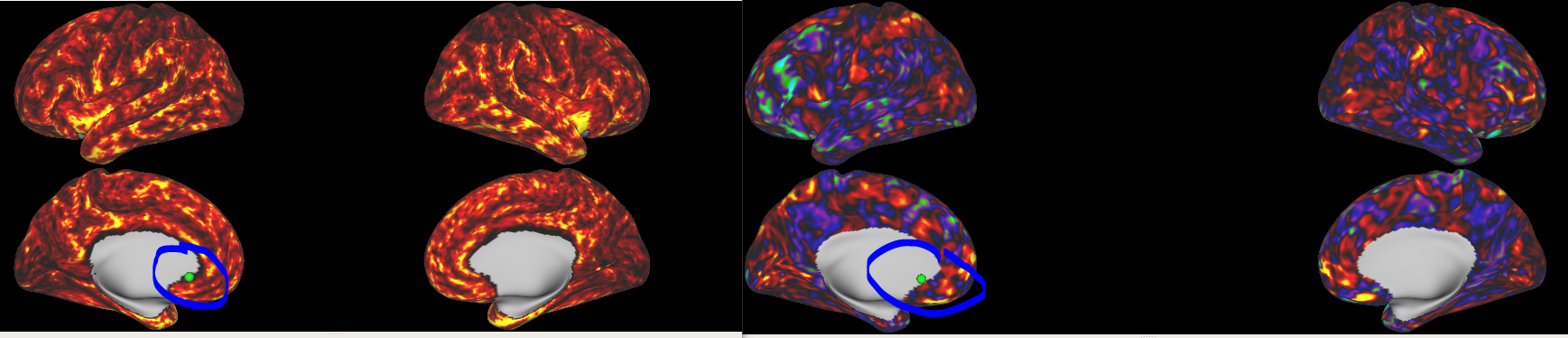
Finally, there are my results, the left is the result of our cortical thickness, and the right is the result of fmriprep’s BOLD preprocessing. They look nice, but our result seems to have eroded a few vertices towards the medial wall. Should we just ignore these vertices? Or could you provide the more specific commands of fmriprep?
I also started a new topic here, https://neurostars.org/t/how-to-extract-cortical-thickness-from-fsaverage-space-to-fslr-den-91k-sapce/20655, providing more details. Thanks a lot!
At first glance, that looks correct - the only suggestion I would have is if you are comparing with the CIFTI surfaces, you will want to ensure the surfaces share # of vertices.
After -metric-resampling, the fsLR surfaces are cleaned to remove any values within the medial wall. This process happens within the CIFTI generation step (you can leverage the tpl-fsLR_hemi-?_den-32k_desc-nomedialwall_dparc.label.gii files for this)
At first glance, that looks correct - the only suggestion I would have is if you are comparing with the CIFTI surfaces, you will want to ensure the surfaces share # of vertices.
After
-metric-resampling, the fsLR surfaces are cleaned to remove any values within the medial wall. This process happens within the CIFTI generation step (you can leverage thetpl-fsLR_hemi-?_den-32k_desc-nomedialwall_dparc.label.giifiles for this)
Thanks for your suggestions! As you said,I want to ensure the surfaces share # of vertices. Please allow me to repeat your words here; our result should be correct but should remove the vertices within the medial wall because those BOLD files did it. But this code CIFTI generation step is difficult for me, could you give me some examples or an alternative "wb_command"?
An alternative way for I compared two files (e.g., thickness and BOLD files), is that I make two files have the same # of vertices based on the bool indices of "tpl-fsLR_hemi-?_den-32k_desc-nomedialwall_dparc.label.gii" . For example, I just extract the 59412 values from all 64984 values based on the vertices position of the nonmedial wall. I think this method also works. Am I right?
However, based on that alternative way, I found something confusing. I had extracted 59412 vertex values from our results, and also read the fmriprep's fsLR32KBOLD file. I think the positions of their non-zero vertices are the same. But our results have more zeros vertices (usually dozens of vertices for the whole cortex) than BOLD. What I chose is the cortical thickness file, does the thickness of the cortex have regions of zero? Besides, for different subjects, their fmriprep's BOLD files have the same vertices of nonzero, but our results do not. So is this situation reasonable? Or we can use "wb_command -cifti-dilate" to fill the zero values near the medial wall, as you mentioned previously in this topic.
This is a late bump, but I'm having a similar issue to the original post with zeroes near the medial wall, across several datasets I'm processing on fMRIPrep 22.0.1 with --cifti-output
Looks like the zeroes are introduced by the fsaverage cortex label masking in mri_vol2surf, (nipype.interfaces.freesurfer.SampleToSurface with cortex_mask=True), and the 32k-resampled BOLD can have zeroed-out vertices that lie outside the fsLR32k medial wall label, which become zeroes in the dtseries output.
Should cortex_mask in SampleToSurface be disabled (or at least have the option to disable) when running with --cifti-output? If I'm not mistaken that should avoid creating the zeroes, while still masking the medial wall on the 32k surfaces.
Hello! I'm seeing this same behavior in a dataset processed with v23.1.4. Can you clarify the following:
- Are these 0-value vertices around the medial wall in the fsLR den-91k dtseries the intended/expected behavior of fMRIprep?
- Is there any way to avoid this and ensure that all vertices outside of the medial wall are populated with data by fMRIprep? Or is -cifti-dilate your recommended solution for those of us unhappy with the 0s?
I'm used to working with CIFTI files where (with rare exceptions) all vertices outside the medial wall contain data. If they don't, it causes headaches down the line to ensure that the 0s don't muck things up when smoothing, parcellating, etc.
Hi @droediger, thanks for posting on this - we hoped #2879 would address this, but it seems there are still cases (potentially due to imperfect segmentations?) where these zeroes can trickle in. Running the dilation step (which is what HCPPipelines is doing) should clean it up.
wb_command -cifti-dilate <fmriprep-dtseries.nii> COLUMN 0 30 <new-dtseries.nii>
Thanks for the reply, @mgxd !
To clarify, it's not just a few cases for me. Every single subject in my current dataset has ~400-600 zero-value vertices in the fsLR dtseries. I'll look more closely, but I didn't notice any obvious issues with the segmentations.
Hi @droediger, thanks for posting on this - we hoped #2879 would address this, but it seems there are still cases (potentially due to imperfect segmentations?) where these zeroes can trickle in. Running the dilation step (which is what HCPPipelines is doing) should clean it up.
wb_command -cifti-dilate <fmriprep-dtseries.nii> COLUMN 0 30 <new-dtseries.nii>
PS A couple questions about the -cifti-dilate command you provided. It's giving an error message that "left surface required but not provided." I'm guessing I should provide midthickness surfaces from the anat folder with the -left-surface and -right-surface args? Secondly, I notice the surface-distance arg is set to 0mm while volume-distance is set to 30mm. Don't we want to dilate on the surface to get rid of the zeroes on the medial wall?
Thanks again, @mgxd !
EDIT: I should also mention that this is a multiecho acquisition and we're using the tedana workflow.
Ah yes, the HCP snippet I linked was for subcortical processing. IIRC, we were hesitant to add dilation because it could potentially mask a problem in the implementation. Processing some (single echo) data with 23.1.4, I was able to replicate your issue, so will start digging and see if this can be tracked down.