SSD decoding example
Reference issue
Fixes #9116.
What does this implement/fix?
An example of how mne.decoding.ssd can be used for enhancing decoding performance. A motor imagery dataset is used. Two examples are implemented, showing how SSD can be applied before data epoching, i.e. outside the decoding pipeline, and after data epoching, i.e. within the decoding pipeline. A note on how SSD can also improve the neurophysiological interpretation of the solution is done at the end of this example.
In addition, since SSD was missing at the tutorials/machine-learning/plot_sensors_decoding.py, a section briefly describing SSD was added.
Additional information
I have my personal concerns regarding the use of SSD before data epoching. Although SSD is an unsupervised spatial filtering approach, there are some parameters that are learned during its implementation. I followed what I understood was done here. Let me know what are your thoughts, I might be missing something.
I open this PR as Draft
For some weird reason, the documentation cannot be built here although my computer is working. It says something regarding memory. Any idea @drammock ???
For some weird reason, the documentation cannot be built here although my computer is working. It says something regarding memory. Any idea @drammock ???
You can run locally as PATTERN=plot_decoding_ssd_csp_eeg make html_dev-pattern-memory to find out how much time/RAM it takes to run the example. I can never remember what the limit is for CircleCI (@larsoner knows I'm sure)... I do know that the limit for building merged PRs is even lower than for open / work-in-progress PRs so some memory optimization is definitely needed.
The rough limit is < 1.5 GB for running one example while building locally. < 1 GB is even better. There are usually ways to trim an example to get to this point via some combination of:
- Cropping raw
- Resampling raw or decimating epochs
- Using fewer epochs
- Picking fewer channels to process
without losing the point of the example. If you try some of these but can't fix things, if you can get the example to a state where it works on your machine and makes the point(s) it should make, I or @drammock could try the tricks we know to trim it and push a commit once the mem usage is down.
The rough limit is < 1.5 GB for running one example while building locally. < 1 GB is even better. There are usually ways to trim an example to get to this point via some combination of:
- Cropping raw
- Resampling raw or decimating epochs
- Using fewer epochs
- Picking fewer channels to process
without losing the point of the example. If you try some of these but can't fix things, if you can get the example to a state where it works on your machine and makes the point(s) it should make, I or @drammock could try the tricks we know to trim it and push a commit once the mem usage is down.
Great, thank you!!! I will do that, I think the for analyzing the impact in n_components in SSD can be trimmer to the half
For some weird reason, the documentation cannot be built here although my computer is working. It says something regarding memory. Any idea @drammock ???
You can run locally as
PATTERN=plot_decoding_ssd_csp_eeg make html_dev-pattern-memoryto find out how much time/RAM it takes to run the example. I can never remember what the limit is for CircleCI (@larsoner knows I'm sure)... I do know that the limit for building merged PRs is even lower than for open / work-in-progress PRs so some memory optimization is definitely needed.
I check this and the memory was 515.7 MB ...so it should work, right?
here are the rendered doc pages:
https://26474-1301584-gh.circle-artifacts.com/0/dev/auto_tutorials/machine-learning/plot_sensors_decoding.html https://26474-1301584-gh.circle-artifacts.com/0/dev/auto_examples/decoding/plot_ssd_spatial_filters.html#sphx-glr-auto-examples-decoding-plot-ssd-spatial-filters-py https://26474-1301584-gh.circle-artifacts.com/0/dev/auto_examples/decoding/plot_decoding_ssd_csp_eeg.html#sphx-glr-auto-examples-decoding-plot-decoding-ssd-csp-eeg-py
I don't know why there is no plot in the second one.
I also find that the error bars are quite big.
thoughts?
here are the rendered doc pages:
https://26474-1301584-gh.circle-artifacts.com/0/dev/auto_tutorials/machine-learning/plot_sensors_decoding.html https://26474-1301584-gh.circle-artifacts.com/0/dev/auto_examples/decoding/plot_ssd_spatial_filters.html#sphx-glr-auto-examples-decoding-plot-ssd-spatial-filters-py https://26474-1301584-gh.circle-artifacts.com/0/dev/auto_examples/decoding/plot_decoding_ssd_csp_eeg.html#sphx-glr-auto-examples-decoding-plot-decoding-ssd-csp-eeg-py
I don't know why there is no plot in the second one.
I also find that the error bars are quite big.
thoughts?
thank you for posting the docs files, I forgot to do it yesterday. The STD is also big for CSP. The reason why that could be happening is because this dataset has only 45 trials. Then, I am taking in the CV the 20% for testing (i.e. only 9 trials are held out). I can try other configurations, but then we have the risk of overfitting. I will check anyways
@dengemann here is the example
sorry I did not look carefully but it is enough to have these 45 trials to make your point here experimentally? I am convinced in theory
I did a pass on the text and comments and made a few suggestions.
thank you @britta-wstnr !
sorry I did not look carefully but it is enough to have these 45 trials to make your point here experimentally? I am convinced in theory …
In theory, yes, using 45 trials should be enough for training a 2 classes problem, but for the practical viewpoint, we might be tight. In order to reduce variance, I have increased the number of random splits from 5 to 10.
Hi ! I have modified the decoding example following your suggestions/comments. Here you can find the new version of it. Thank you!
I am not sure I agree with the statements in this tutorial.
When I see https://26588-1301584-gh.circle-artifacts.com/0/dev/_images/sphx_glr_plot_decoding_ssd_csp_eeg_001.png I see that SSD improves a bit when you overfit by using the full data (SSD outside of the pipeline).
When you do it inside the pipeline https://26588-1301584-gh.circle-artifacts.com/0/dev/_images/sphx_glr_plot_decoding_ssd_csp_eeg_002.png I see no reall difference between CSP and SSD+CSP.
to me the take home message of this example should be "do not do SSD outside of the pipeline it is not statistically OK"
I am not sure I agree with the statements in this tutorial. When I see https://26588-1301584-gh.circle-artifacts.com/0/dev/_images/sphx_glr_plot_decoding_ssd_csp_eeg_001.png I see that SSD improves a bit when you overfit by using the full data (SSD outside of the pipeline). When you do it inside the pipeline https://26588-1301584-gh.circle-artifacts.com/0/dev/_images/sphx_glr_plot_decoding_ssd_csp_eeg_002.png I see no reall difference between CSP and SSD+CSP. to me the take home message of this example should be "do not do SSD outside of the pipeline it is not statistically OK"
I totally understand your concern @agramfort , which in fact are my concerns too. Although I personally think that all pre-processing methods should be considered within the pipeline, to avoid overfitting, this was not the intended main message of this tutorial. So, if this is what is being caught, we can just avoid the use of SSD before data epoching and try to find a better way to show the potential of SSD when used within the pipeline?
I totally understand your concern @agramfort https://github.com/agramfort , which in fact are my concerns too. Although I personally think that all pre-processing methods should be considered within the pipeline, to avoid overfitting, this was not the intended main message of this tutorial. So, if this is what is being caught, we can just avoid the use of SSD before data epoching and try to find a better way to show the potential of SSD when used within the pipeline?
+1 to consider this unconvincing. Ok for you?
I totally understand your concern @agramfort https://github.com/agramfort , which in fact are my concerns too. Although I personally think that all pre-processing methods should be considered within the pipeline, to avoid overfitting, this was not the intended main message of this tutorial. So, if this is what is being caught, we can just avoid the use of SSD before data epoching and try to find a better way to show the potential of SSD when used within the pipeline? +1 to consider this unconvincing. Ok for you?
This morning I realized that the window size was too small for running SSD within the pipeline. I haven't finalized yet the new example version, but this is the output I got now. Are you suggesting closing this PR or not including the example of SSD outside the pipeline?
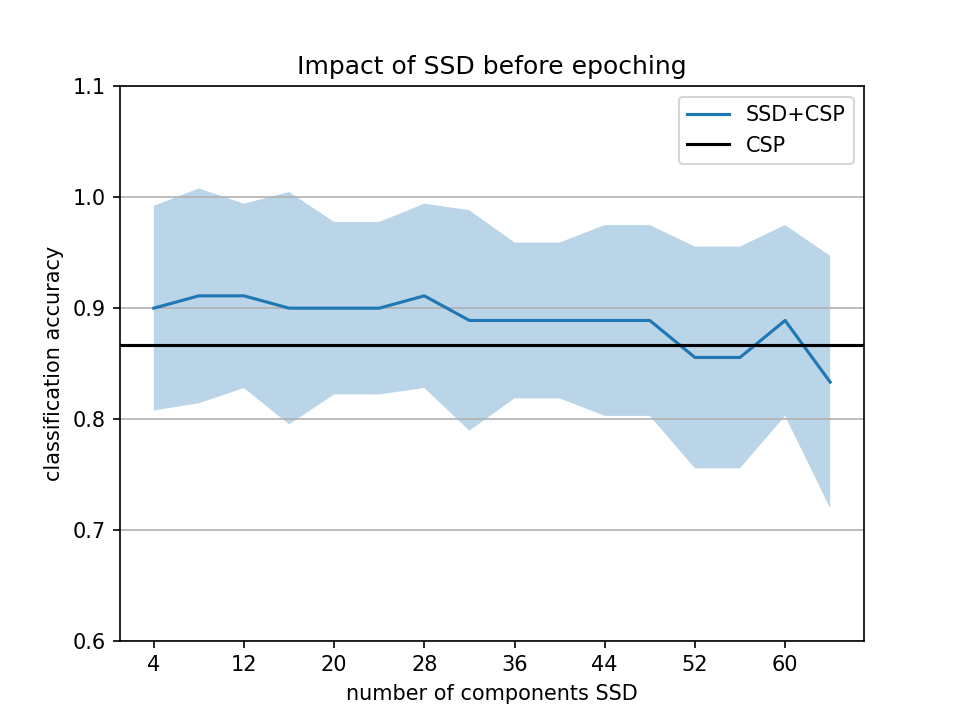
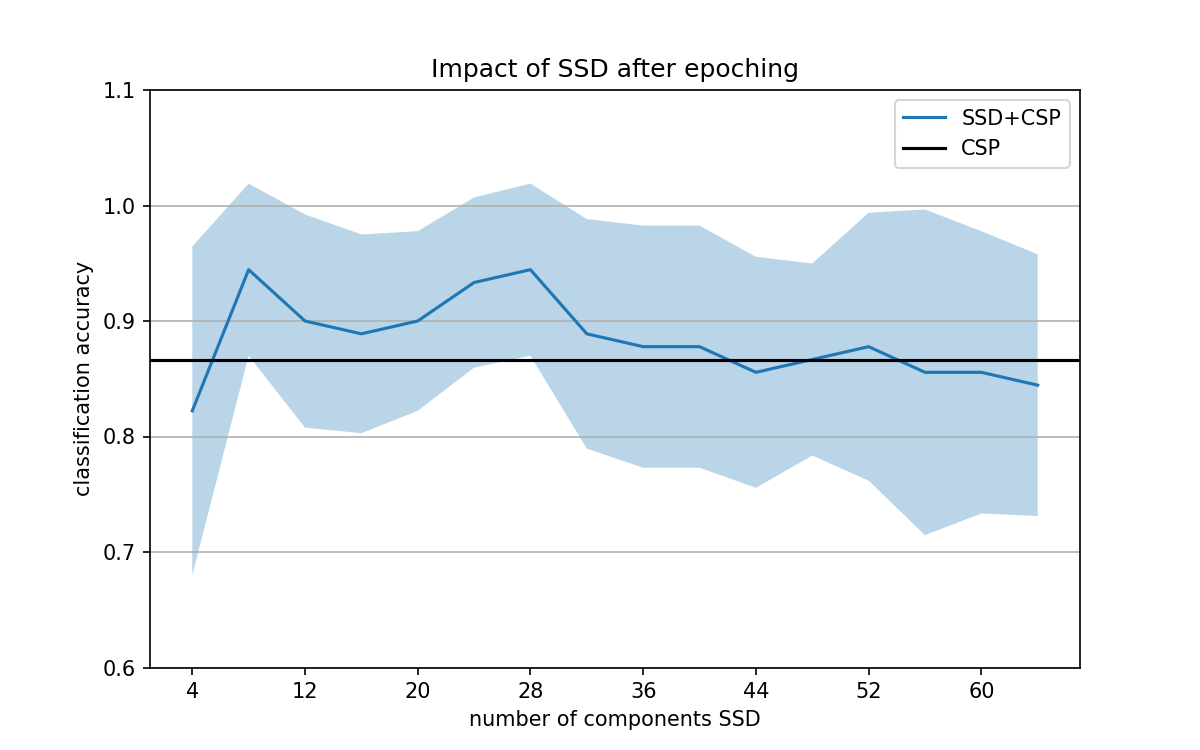
if we run this for more subjects and average can it be even clearer? I find it still not super obvious sorry
if we run this for more subjects and average can it be even clearer? I find it still not super obvious sorry …
Sure, I can do that, it might take more memory but hopefully, the docfiles would be able to be compiled anyway. Anyhow, in the best-case scenario, we would achieve something similar to Figure 4b of Haufe et al. paper. I think that the last attached Figure (SSD after epoching) is, more or less, already in that shape, isn't it?. I can also connect tomorrow to Discord to further discuss this if you are available @agramfort ! thank you!
yes exactly like this figure. I see it's obtained by averaging 80 subjects. Let's see if you can replicate this figure without limitation of computation time on your machine and then we try to find a compromise on the example
ok?
Hi @agramfort. I have run the script over the first 100 subjects available in the dataset. Then, I picked up those subjects whose decoding performance was around the same value (to avoid large std values due to differences in performance). This is what I got for those 10 subjects when SSD is applied after epoching.
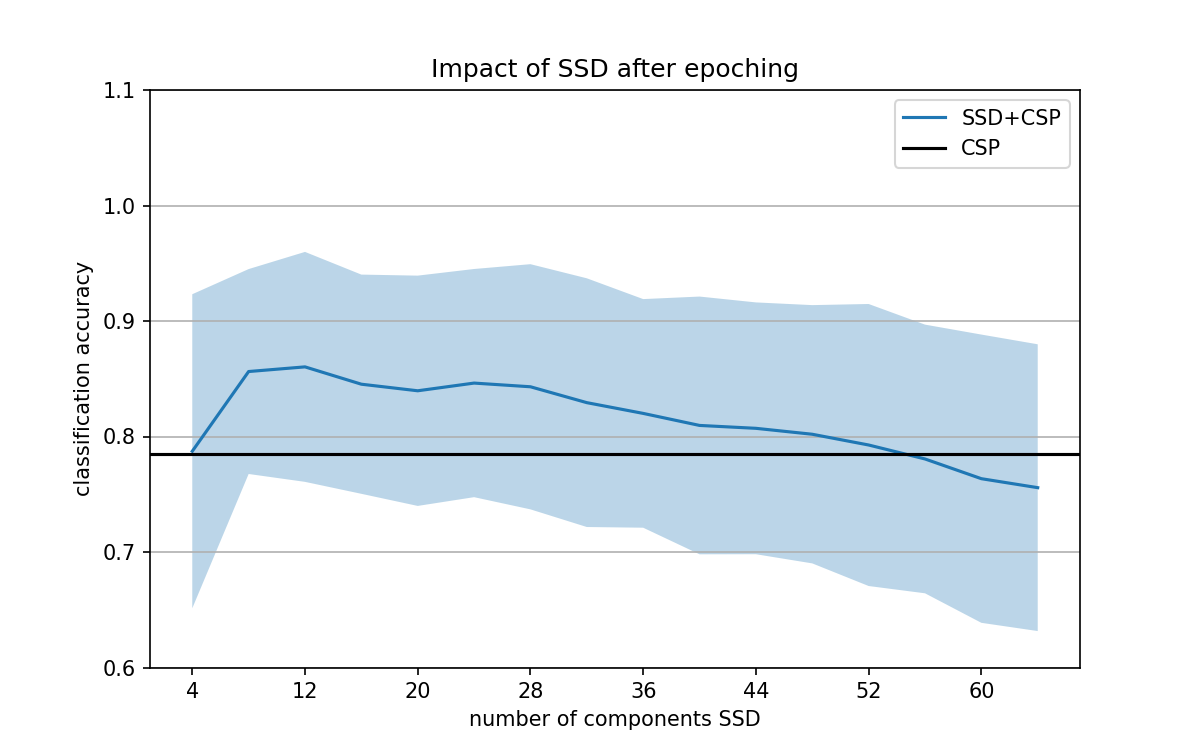
I think these results better show now that SSD can be used before decoding for improving performance. As also reported by the authors, overall classification performance reaches its maximum for a small SSD number of components (here between 6 and 12), and then classification decreases as the number of components increases.
Also, for one of those subjects, I plot the topographical map of the CSP patterns. This is how the patterns look without SSD
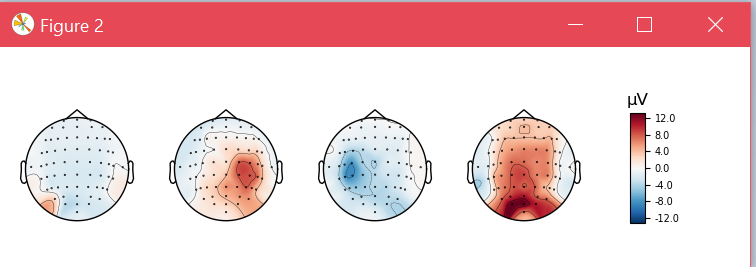
and these are the CSP patterns for the same subject when SSD was applied before learning CSP.
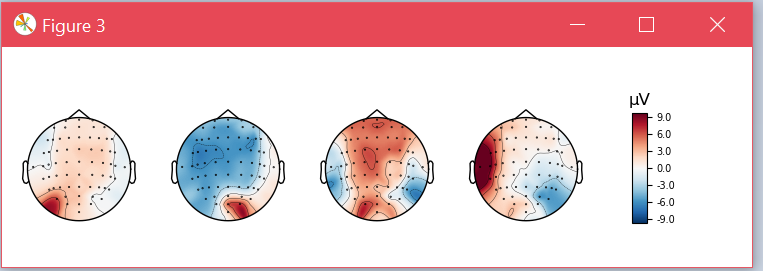
in which, since the experiment is done base on the alpha band, we can see how the patterns now better enhance the occipital region (which could be more related to the visual stimulus rather than the MI process)
Let me know if this makes more sense now. Anyhow, I understand if you consider this example is not strong enough and the PR should then be closed.
Thank you,
this was obtained using a proper pipeline? without overfitting? if so you would update the tutorial demoing this? how long does the example take to run?
this was obtained using a proper pipeline? without overfitting? if so you would update the tutorial demoing this? how long does the example take to run? …
yes, it is obtained with the non-overfitting pipeline. But on my computer takes 20 minutes to run everything. I understand that is not doable for online docfiles creation, right? I will commit the changes anyway so as to keep track of that here too.
we do have 1 or 2 tutorials that we simply show the code but don't execute them during our doc build. This is achieved by naming the tutoral something.py instead of plot_something.py. Is that a suitable option here @agramfort? Or is the result of the 20-minute build still not convincing enough to be worth showcasing?