mne-bids-pipeline
 mne-bids-pipeline copied to clipboard
mne-bids-pipeline copied to clipboard
Decoding in time-frequency space using CSP + Permutation statistics
This PR adds a new script to the pipeline : 03b_time_frequency_csp.py, designed to analyze in the time-frequency domain a contrast between two conditions.
There are two main steps in this script:
- Decoding for each time-frequency bin: For each time-frequency bin, we use a csp classifier in order to distinguish between the two conditions. We compute the roc-auc score for each time-frequency bin.
- Permutation statistics at the group level: We try to answer the following question: is the difference between the two conditions statistically significant? We use the classic permutations cluster tests on the time-frequency roc-auc map.
During the script, we create a new type of reports: *-proc-csp+permutation+test_report.html, that contain the following figures :
-
Frequency roc-auc decoding:
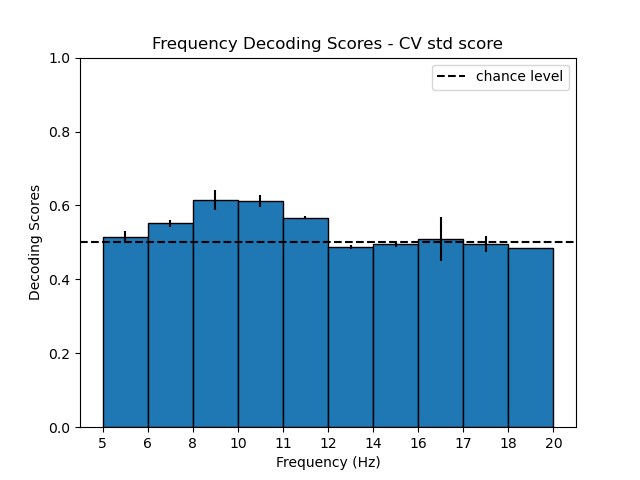
-
Time-frequency decoding
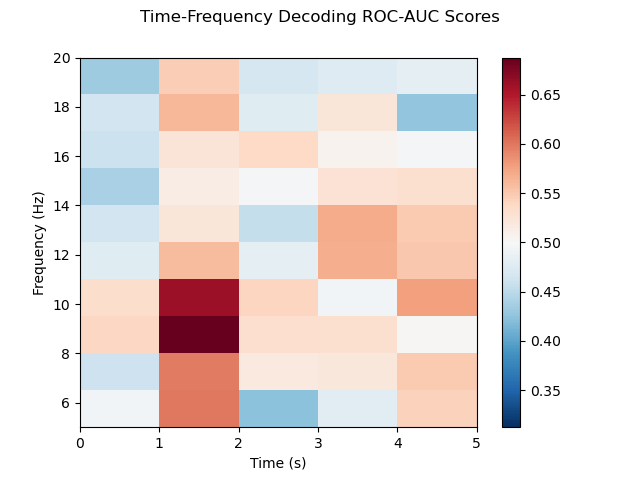
-
Permutation statistics
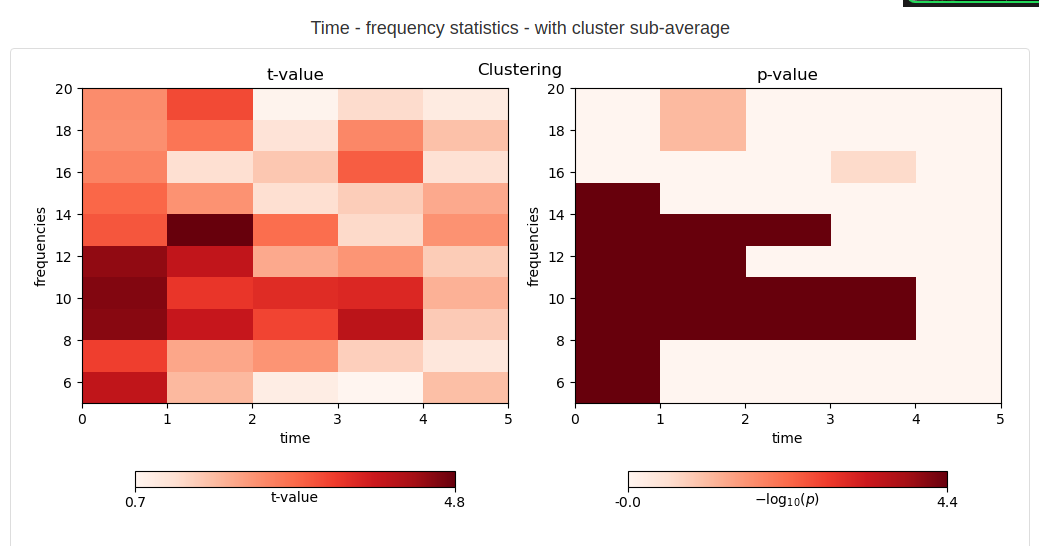
CI Test: WIP The code was tested in the working memory time study with 20 subjects. [The code also passed the CI tests, although the majority of the CI test is not designed to test this type of code, specifically designed for group studies. WIP]
WIP:
- [x] Change log entry
- [x] Run time optimization
- [x] Reports
Coding style: In the code, I use two utils class : Tf, and Pth, each one dealing with time-frequency useful information, and paths management. This is a bit of a disruption because there are no such class in the rest of the pipeline, but they have enabled to DRY up the code significantly.
Thanks @crsegerie 🚀 I will review tomorrow morning 👌
First small round of reviews, more to come :)
Green, finally !
Tests details:
| name | Nsubjects | Nruns | Ncontrasts | ch_types | CSP | CSP Warning |
|---|---|---|---|---|---|---|
| config_ds000117 | 1 | 2 | 3 | ['meg'] | Small warning because of time ordering + Warning because decimation | |
| config_ds000246 | 1 | 1 | 1 | ['meg'] | MAG dimension 247 reduced manually to 100. | |
| config_ds000247 | 1 | Nan | Nan | ['meg'] | Contrast = 0 so CSP not run | |
| config_ds000248 | 1 | Nan | 2 | ['meg'] | ||
| config_ds000248_coreg_surfaces | 1 | Nan | Nan | ['meg'] | Contrast = 0 so CSP not run | |
| config_ds000248_FLASH_BEM | 1 | Nan | Nan | ['meg'] | Contrast = 0 so CSP not run | |
| config_ds000248_ica | 1 | Nan | Nan | ['meg'] | Contrast = 0 so CSP not run | |
| config_ds000248_no_mri | 1 | Nan | 1 | ['meg'] | ||
| config_ds000248_T1_BEM | 1 | Nan | Nan | ['meg'] | Contrast = 0 so CSP not run | |
| config_ds001810 | 5 | Nan | 1 | ['eeg'] | (Small warning because of decimation, otherwise perfect) | |
| config_ds001971 | 1 | 1 | 1 | ['eeg'] | ||
| config_ds003104 | Nan | Nan | Nan | ['meg'] | Contrast = 0 so CSP not run | |
| config_ds003392 | 1 | Nan | 1 | ['meg'] | ||
| config_eeg_matchingpennies | 1 | Nan | 1 | ['eeg'] |
I made a small script to extract into a csv the configuration settings to check that the new script has been tested in all major configurations.
Runtime: Overall, even though the new script would take a long time to compute naively, with proper decimation and dimension reduction, the script runs in less than ~30 seconds for all tests.
It's not perfect yet, but I think I can hand the baton over to you. I hope this script won't contain too many anti-patterns, and that it won't be too hard to merge... but before the end of my internship, I have to move on to other things.
I'm glad I've been able to work on it. Thank you @agramfort for the various optimization tips, thank you @hoechenberger for the peer coding sessions, thank you @SophieHerbst for the study idea!