mne-bids-pipeline
 mne-bids-pipeline copied to clipboard
mne-bids-pipeline copied to clipboard
Improve Reports
The generated reports currently leave much to be desired. I will update this list as I discover more missing features / issues, and of course tick them off if issues get resolved :)
- [x] there's no Epochs drop log (I believe we used to have one, once upon a time?) (https://github.com/mne-tools/mne-python/pull/7990)
- [x] PSD plots for raw data missing (#155)
- [x] SSP projectors are missing (https://github.com/mne-tools/mne-python/pull/7991)
- [x] decoding results are not included (#184)
- [ ] TFR results are not included
- [x] I don't think we're currently including diagnostic info re ICA (#163)
- [x] some results appear twice, because we're using
parse_folder()and later adding these elements again, manually - [x] the buttons in the top-right toggle visibility of sections, however I suppose most users would assume they're navigation links, so their behavior is confusing
Some of these issues should be fixed upstream?
Some of these issues should be fixed upstream?
Absolutely, I’m just trying to bundle things here a bit to have this list in one place and not split across a few :)
I think this might be useful to note here...
I have a dataset with N=37 subjects. The sub-Average report shows me a subject trial counts which is brilliant. But all the plots and accompanying .mat files--why .mat files? 😩--lead me to believe the evoked file is for a single subject...please say it's not so.
Maybe an auxiliary .tsv for subjects in sub-Average. Just a thought.
On second thought, I am not sure if the Events counts is very informative, since in all cases trial counts should be equalized, I think a dot plot of subjects x counts x events LUT. Saves space too...just food for thought.
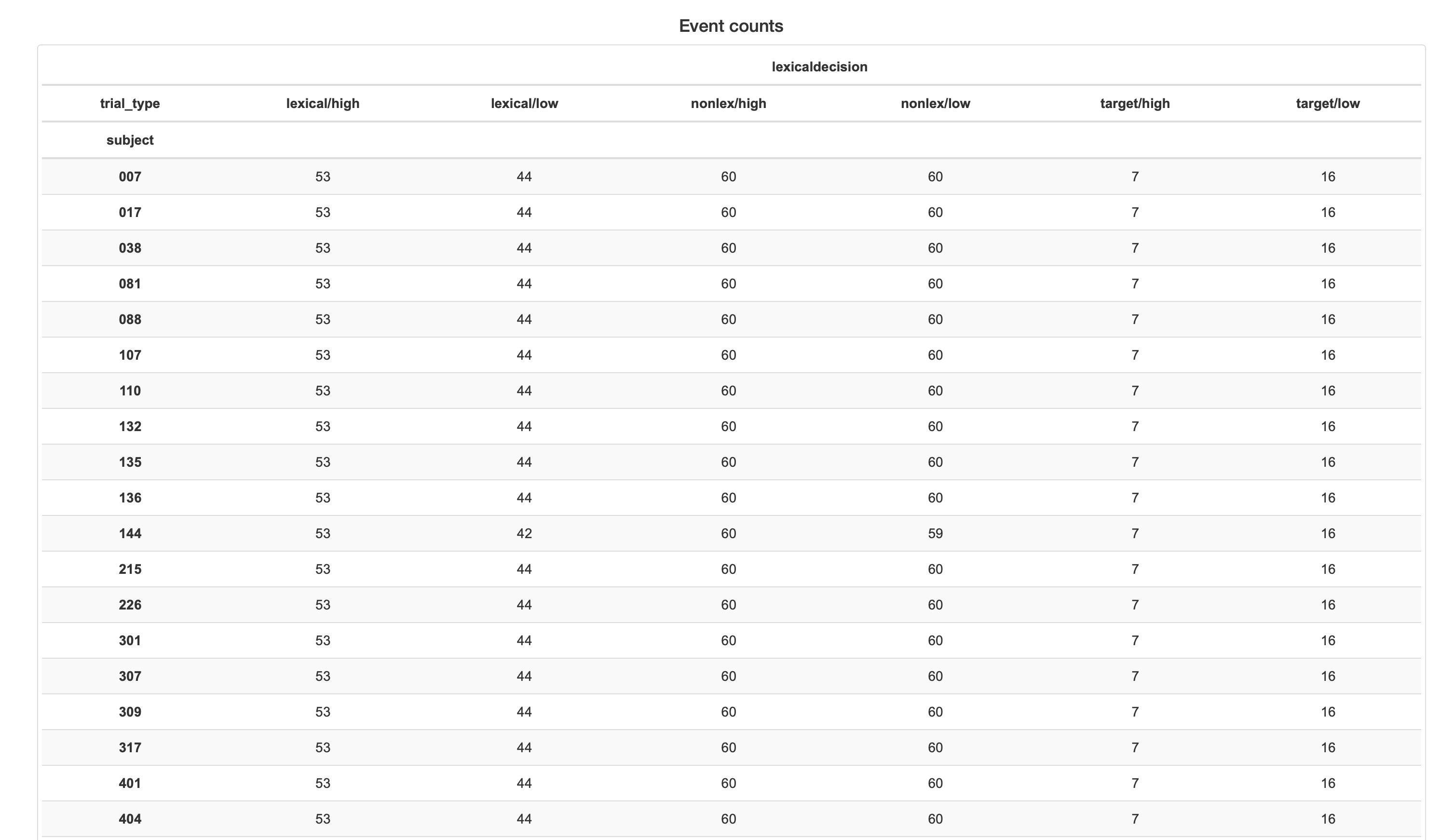
Upstream issue with legend and topo map positions?
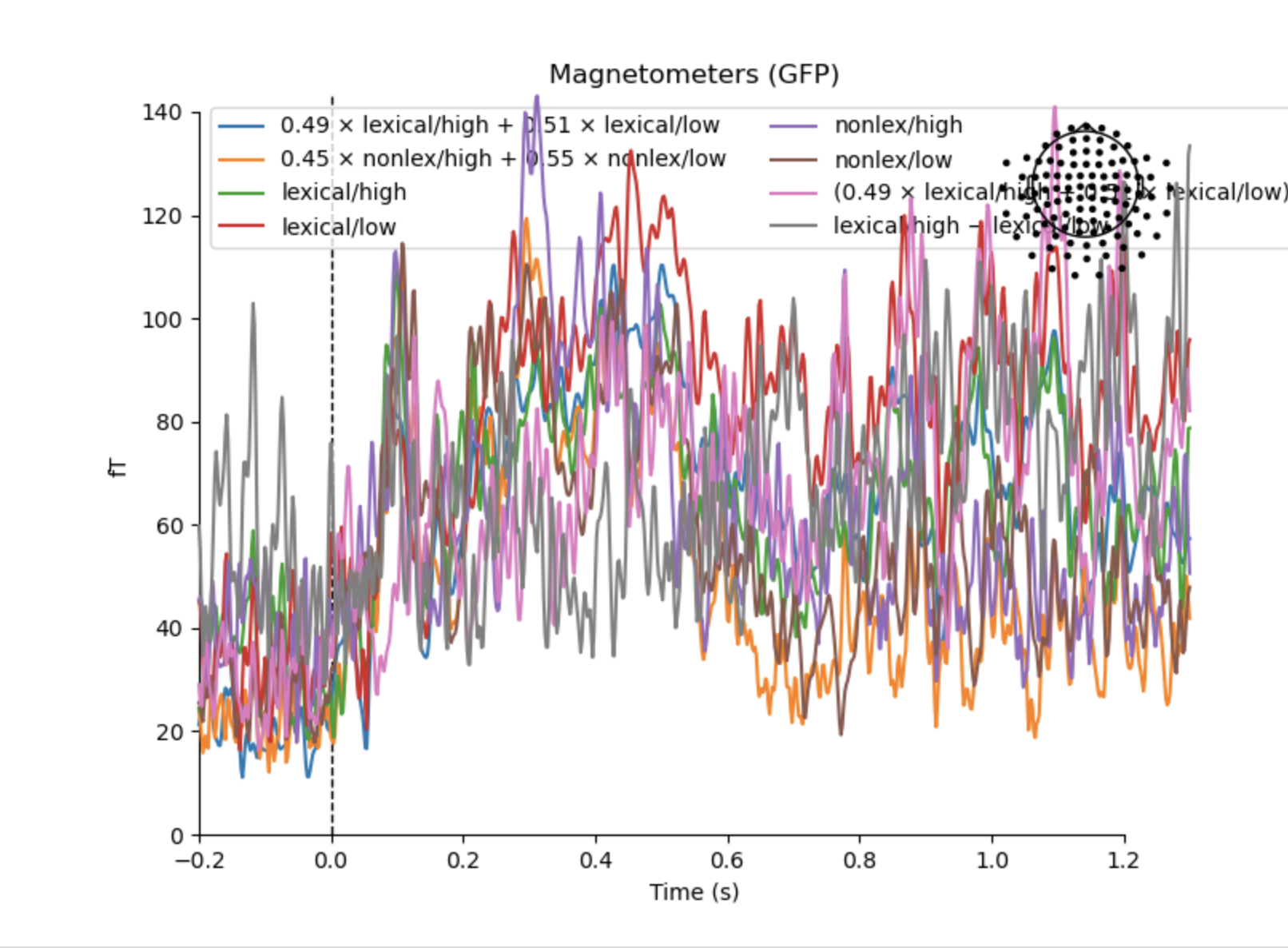
we use .mat files for things that cannot be stored in .fif (eg decoding scores).
your decoding plot seems just for one subject.
Upstream issue with legend and topo map positions?
Yeah, and the big legend is clearly a problem. We'll need to address this upstream, or revise this plot entirely somehow – there's just too many conditions here to make anything useful out of it anyway, no? Any suggestions on how we could improve things?
On second thought, I am not sure if the Events counts is very informative, since in all cases trial counts should be equalized, I think a dot plot of subjects x counts x events LUT. Saves space too...just food for thought.
So this table is actually only based on the data included in the unprocessed dataset and doesn't take into account any sorts of cleaning / epochs rejection etc. That's definitely something we need to address. The table was originally implemented outside of the study template in MNE-BIDS to help users get a quick idea about what's contained in a specific dataset. If anything, we should label it accordingly: "This is what we found in the dataset before any data cleaning". But ideally we'd have something the reflects the data that actually made it into the analysis results!
What is "LUT"?
But all the plots and accompanying
.matfiles--why.matfiles? 😩--lead me to believe the evoked file is for a single subject...please say it's not so.
I'm not sure I can exactly follow you here … inside the sub-XXX directories, we'd always only store processing results for that specific participant; so any epochs, evoked, ... file in there will always only pertain to that very participant. The sub-average contains the data averaged across participants.
Are you suggesting we also supply an evoked file that contains a list of evokeds, where each element would correspond to individual participants? If yes, I believe we'd have to create separate files for each condition to allow us to properly categorize things. Would that address your need & usecase?
Thanks!
why
.matfiles?
I believe these are actually just HDF5 files these days :) Any suggestions for a different data format that would make your life easier? Is it even clear what these files are supposed to contain? (Don't get me wrong – not questioning your cognitive abilities ;) merely wondering if and how we should improve documentation!!) Thank you!
your decoding plot seems just for one subject.
@agramfort that's what I am thinking. Considering it's from the sub-average report I suspect something went wrong.
Yeah, and the big legend is clearly a problem. We'll need to address this upstream, or revise this plot entirely somehow – there's just too many conditions here to make anything useful out of it anyway, no? Any suggestions on how we could improve things?
Actually I only have four event types; I am not really sure what the X x A + Y x B evoked conditions represent. Also, considering this is a GFP plot, the sensor map seems unnecessary.
So this table is actually only based on the data included in the unprocessed dataset and doesn't take into account any sorts of cleaning / epochs rejection etc.
The individual subject reports show event counts using an event_id x time with arbitrary? color markers for id. Perhaps this can be adapted into an id x subject plot using color/size semantics
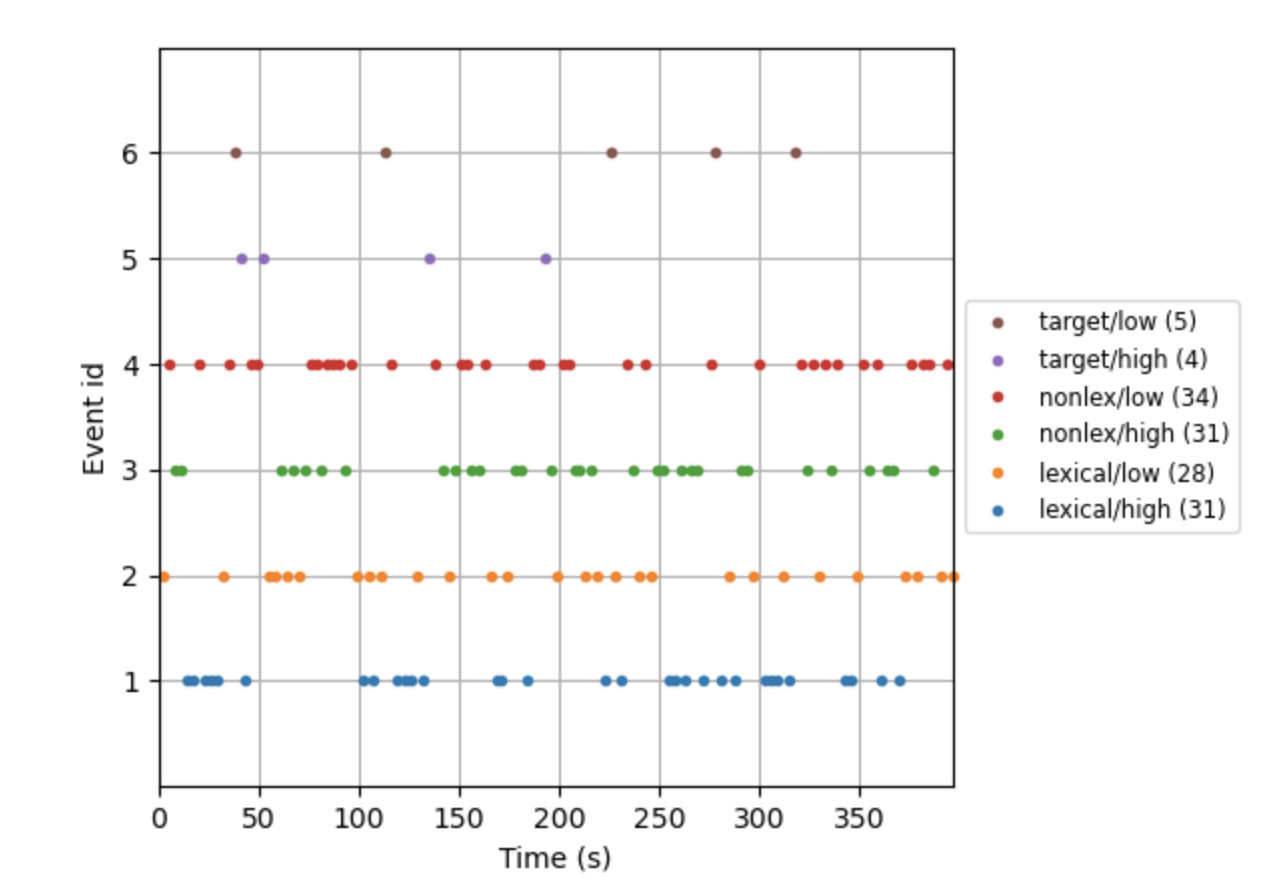
Are you suggesting we also supply an evoked file that contains a list of evokeds, where each element would correspond to individual participants? If yes, I believe we'd have to create separate files for each condition to allow us to properly categorize things. Would that address your need & usecase?
I guess the question is whether BIDS specifies information on data that went into sub-Average e.g. json or tsv listing individual data used to compute averaging. I imagine this becomes useful if not necessary in the case of source imaging where parameters differ from sensor level routines.
Any suggestions for a different data format that would make your life easier?
csv plays nice with other APIs and stats software.
Is it even clear what these files are supposed to contain? (Don't get me wrong – not questioning your cognitive abilities ;) merely wondering if and how we should improve documentation!!)
I am in the basement level of Maslow's hierarchy so my cognition is pretty rudimentary these days :)) I noticed HD5 files in my single subject directory(s). But I presume @agramfort the decoding scores will in most cases be combined across subjects for a particular study for illustrative purposes, so if you're looking to make my life easier @hoechenberger then either HD5, CSV --or I've been known to dabble in XARRAY.
or I've been known to dabble in XARRAY.
I'm a big fan of xarray too, WDYT about that, @agramfort?
sorry what is the question?
sorry what is the question?
Whether it could make sense to replace the .mat files we currently produce with something else. @ktavabi suggested xarray could make sense.
good idea !
xarray allows for various storage options, including netCDF; but one caught my attention: they have a dictionary export that would then allow one to store the data in a JSON file. WDYT about that?
Personally I would just use h5io / mne.externals.h5io since it's a thin layer over hdf5 which can be read most places without issue.
the benefit of xarray is to have index for all axis (time, subject, frequency, cross-val fold etc.) as we don't have proper containers for decoding scores
Personally I would just use h5io / mne.externals.h5io since it's a thin layer over hdf5 which can be read most places without issue.
@larsoner FWIW my impression is that xarray is more versatile than hdf5, as it's developed specifically for working with ND-array datasets contained in numpy or pandas frames, and IIRC can handle dictionary I/o from hdf5 containers.
here is a result with a sub-Average for decoding:
http://mne.tools/mne-bids-pipeline/examples/ERP_CORE/sub-average_ses-N400_task-N400_report.html#5
you don't get the same?
no 😒 I'm beginning to suspect there are pop-up corner cases along the pipeline...
Presently trying to run all steps on entire cohort but via pdb due to critical errors with different individual datasets...
04:31:34 [Step-05][sub-301] Saving epochs Overwriting existing file. 04:31:34 A critical error occurred. Traceback (most recent call last): ...events, mappings = _read_events_fif(fid, tree) File "/Users/ktavabi/Github/mne-python/mne/event.py", line 167, in _read_events_fif event_list.shape = (-1, 3) AttributeError: 'NoneType' object has no attribute 'shape'
...
11:46:35 [Step-07][sub-215] Contrasting conditions: lexical/high – lexical/low 11:46:35 A critical error occurred. Traceback (most recent call last): ... ValueError: n_splits=5 cannot be greater than the number of members in each class. /Users/ktavabi/opt/miniconda3/envs/obiwan/lib/python3.9/site-packages/sklearn/model_selection/_split.py(662)_make_test_folds() raise ValueError("n_splits=%d cannot be greater than the"
Probably irrelevant here now.
FWIW I've previously analyzed this data set using my caveman scripts, using Autoreject for epoching. It looks like all my post preprocessing runtime errors are downstream from trial scoring.
did you check the _events.tsv files? they look ok?
the bids validator is happy with your dataset?
did you check the _events.tsv files?
I will do that this weekend for the problematic cases.
the bids validator is happy with your dataset?
Yes, sir! men-bids ran through without error
04:31:34 [Step-05][sub-301] Saving epochs Overwriting existing file. 04:31:34 A critical error occurred. Traceback (most recent call last): ...events, mappings = _read_events_fif(fid, tree) File "/Users/ktavabi/Github/mne-python/mne/event.py", line 167, in _read_events_fif event_list.shape = (-1, 3) AttributeError: 'NoneType' object has no attribute 'shape'
...
11:46:35 [Step-07][sub-215] Contrasting conditions: lexical/high – lexical/low 11:46:35 A critical error occurred. Traceback (most recent call last): ... ValueError: n_splits=5 cannot be greater than the number of members in each class. /Users/ktavabi/opt/miniconda3/envs/obiwan/lib/python3.9/site-packages/sklearn/model_selection/_split.py(662)_make_test_folds() raise ValueError("n_splits=%d cannot be greater than the"
Wow this looks super weird… You wouldn't be able, by any chance, to share (part of) the problematic dataset?
HERE are the BIDS source and pipeline payloads for accompanying config.
If fixed it in dropbox. One of the subjects had a reject which was too low and leading to zero epochs.
Thanks @agramfort Will try with cohort next. The pipeline doesn't re-run routines like epoching prior to grand averaging, decoding, or TF steps given necessary derivative files are present. Correct?
The pipeline doesn't re-run routines like epoching prior to grand averaging, decoding, or TF steps given necessary derivative files are present. Correct?
Yes this is correct if you request to only run the respective processing steps (i.e., sensor/group_average and report)
If you simply re-run the entire pipeline, it will regenerate everything.
However, if the required derivatives are not present, I believe we'll just crash horribly. 😅
updates look great! FWIW mne_report gives off nice profiler vibes 😄
Good to hear it's working!
Please do feel free to share anything you don't like about the report. There's so many things to be improved there, but user feedback can help us prioritize things.