torch
 torch copied to clipboard
torch copied to clipboard
Weird runtime behaviour
Hi @dfalbel,
While benchmarking some R packages for DL (torch is the fastest) we made a strange observation. The runtime of torch suddenly doubles at a certain network size (the threshold is very specific):
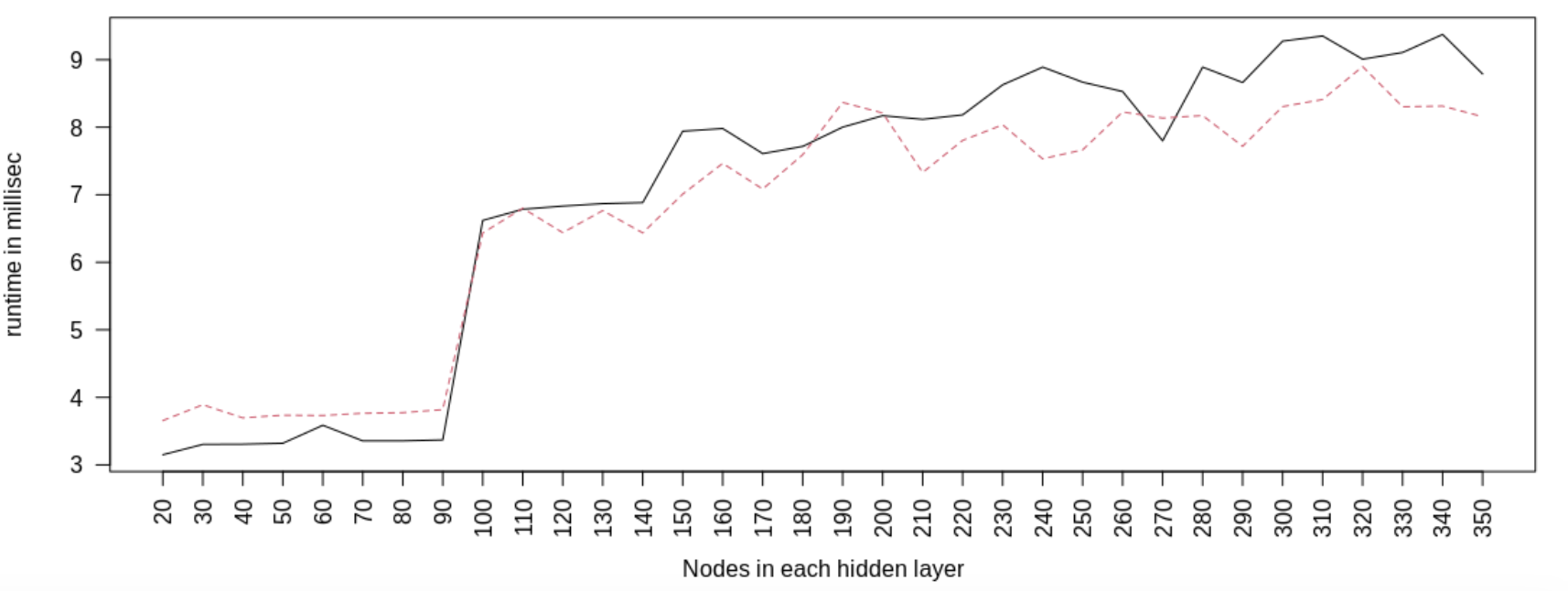
It seems to be caused by the optimizer, because benchmarking the forward function shows that the predictions are in the microsecond range without the sudden runtime step:

The same behaviour appeared on another machine (also Linux), but the position of the step shifted slightly.
Code:
library(torch)
set.seed(42)
torch::torch_manual_seed(42)
X = matrix(runif(20*1000), 1000, 20)
Y = runif(1000)
data = data.frame(X = X, Y = Y)
N = seq(20, 350, by = 10)
loss = nnf_mse_loss
train_func = function(model, optim) {
indices = sample.int(nrow(data), 32L)
XT = torch::torch_tensor(as.matrix(data[indices,-ncol(data)]))
YT = torch::torch_tensor(as.matrix(data[indices,ncol(data)]))
optim$zero_grad()
pred = model$forward(XT)
ll = loss(pred, YT)
ll$backward()
optim$step()
}
res1 =
sapply(N, function(i) {
model = torch::nn_sequential(nn_linear(20L, i), nn_relu(), nn_linear(i, i), nn_relu(), nn_linear(i, 1L))
opt = torch::optim_sgd(params = model$parameters, lr = 0.01)
R = microbenchmark::microbenchmark(train_func(model, opt))
return(summary(R)$median)
})
res2 =
sapply(N, function(i) {
mlp_module <-
torch::nn_module(
"mlp_module",
initialize = function() {
self$model = torch::nn_sequential(nn_linear(20L, i), nn_relu(), nn_linear(i, i), nn_relu(), nn_linear(i, 1L))
},
forward = function(x) {
self$model(x)
}
)
model = mlp_module()
opt = torch::optim_sgd(params = model$parameters, lr = 0.01)
R = microbenchmark::microbenchmark(train_func(model, opt))
return(summary(R)$median)
})
matplot(cbind(res1, res2), type = "l", xaxt = "n", las = 1, xlab = "Nodes in each hidden layer", ylab = "runtime in millisec")
axis(1, at = 1:length(N), labels = N, las = 2)
res_pred =
sapply(N, function(i) {
indices = sample.int(nrow(data), 32L)
XT = torch::torch_tensor(as.matrix(data[indices,-ncol(data)]))
model = torch::nn_sequential(nn_linear(20L, i), nn_relu(), nn_linear(i, i), nn_relu(), nn_linear(i, 1L))
R = microbenchmark::microbenchmark(model(XT))
return(summary(R)$median)
})
plot(res_pred, type = "l", xaxt = "n", las = 1, xlab = "Nodes in each hidden layer", ylab = "runtime in microseconds")
axis(1, at = 1:length(N), labels = N, las = 2)
Session Info:
R version 4.2.2 Patched (2022-11-10 r83330)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.6 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] torch_0.9.1 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.9 purrr_0.3.4 readr_2.1.2
[7] tidyr_1.2.0 tibble_3.1.7 tidyverse_1.3.1 rbenchmark_1.0.0 ggplot2_3.3.6 cito_1.0.1
[13] brulee_0.2.0 neuralnet_1.44.2 h2o_3.38.0.1
loaded via a namespace (and not attached):
[1] httr_1.4.3 bit64_4.0.5 jsonlite_1.8.0 splines_4.2.2 modelr_0.1.8
[6] microbenchmark_1.4.9 assertthat_0.2.1 cellranger_1.1.0 pillar_1.7.0 backports_1.4.1
[11] lattice_0.20-45 glue_1.6.2 checkmate_2.1.0 rvest_1.0.2 hardhat_1.2.0
[16] colorspace_2.0-3 sandwich_3.0-2 Matrix_1.5-1 pkgconfig_2.0.3 broom_1.0.0
[21] haven_2.5.0 mvtnorm_1.1-3 scales_1.2.0 processx_3.6.1 tzdb_0.3.0
[26] generics_0.1.2 ellipsis_0.3.2 TH.data_1.1-1 withr_2.5.0 cli_3.3.0
[31] survival_3.4-0 magrittr_2.0.3 crayon_1.5.1 readxl_1.4.0 ps_1.7.1
[36] fs_1.5.2 fansi_1.0.3 MASS_7.3-58 xml2_1.3.3 tools_4.2.2
[41] data.table_1.14.2 hms_1.1.1 lifecycle_1.0.1 multcomp_1.4-19 munsell_0.5.0
[46] reprex_2.0.1 callr_3.7.0 compiler_4.2.2 rlang_1.0.3 grid_4.2.2
[51] RCurl_1.98-1.9 rstudioapi_0.13 bitops_1.0-7 gtable_0.3.0 codetools_0.2-18
[56] DBI_1.1.3 curl_4.3.2 R6_2.5.1 zoo_1.8-10 lubridate_1.8.0
[61] bit_4.0.4 utf8_1.2.2 coro_1.0.2 stringi_1.7.6 Rcpp_1.0.8.3
[66] vctrs_0.4.1 dbplyr_2.2.1 tidyselect_1.1.2
Hi @MaximilianPi ,
Thanks for the detailed benchmarks! That's nice! Is suspect that starting from that point we are calling GC in every backward iteration and thus adding a large overhead. You could try playing with some of the memory management options described in:
https://torch.mlverse.org/docs/articles/memory-management.html#cpu
If changing the threshold changes the results then that is probably the cause of the large step.