VIBE
 VIBE copied to clipboard
VIBE copied to clipboard
Reproduction of the published results
Hello, unfortunately, I am still not able to reproduce the results published in the paper. According to #98 and #69, this is an issue not only for me but also for many others. I would be happy if someone that tried to reproduce the results can give feedback if they have been able to resolve this issue @shyanguan, @ZephirGe, @Ling-wei, @chenshudong.
Additionally, I tried to analyze the effect of the discriminator, which gave me results unfitting to those reported in the ablation study of the paper.
I will discuss both issues in detail below.
- Regarding the reproduction of the results. I followed the reply of one of the authors @mkocabas in #69:
There are a couple of things we need to be careful when interpreting the results. The results we see on the terminal output or train_log.txt file obtained at the end of each epoch are on the validation set. For the 3DPW dataset, the validation set is harder than the test set, so it is expected to get worse results. You need to run eval.py script to get the results on the test set.
However, running eval.py using my trained model gives me the following result:
MPJPE: 88.1433, PA-MPJPE: 54.4325, PVE: 103.7052, ACCEL: 24.0702, ACCEL_ERR: 25.0214,, which still does not match published performance.
I also tried to verify the claim that the 3DPW validation set is harder than the test set.
Therefore I run the evaluation script using the pre-trained network weights provided by the authors (vibe_model_w_3dpw.pth.tar) on the validation set. Surprisingly this gave me the following result:
MPJPE: 66.1006, PA-MPJPE: 45.5599, PVE: 80.2233, ACCEL: 25.9692, ACCEL_ERR: 27.3321,.
The MPJPE, PA-MPJPE, and PVE are all significantly lower than the respective results from the test set:
MPJPE: 82.9725, PA-MPJPE: 52.0008, PVE: 99.1108, ACCEL: 22.3743, ACCEL_ERR: 23.4263,, and far from any values I can observe during training using the provided code.
For me, this unexpectedly good performance on the validation set seems strange.
And also contradictory to the claim that the validation set is harder than the test set.
- Regarding analyzing the effect of the generator. I trained the model with D_MOTION_LOSS_W set to 0.0 to disable any effect of the discriminator on the generator. To my understanding, this should replicate the results of "Baseline (only G)" presented in Table 2 of the ablation study of the paper.
Quite surprisingly I obtained the following result on the test set of 3DPW:
MPJPE: 85.5106, PA-MPJPE: 53.4883, PVE: 101.1881, ACCEL: 23.9212, ACCEL_ERR: 24.8983,.
This is i) significantly better than the results reported in Table 2 of the ablation study and ii) marginally better than the results I obtain when training with the discriminator.
I repeated both experiments for two times. Corresponding TensorBoard and console logs can be found below.
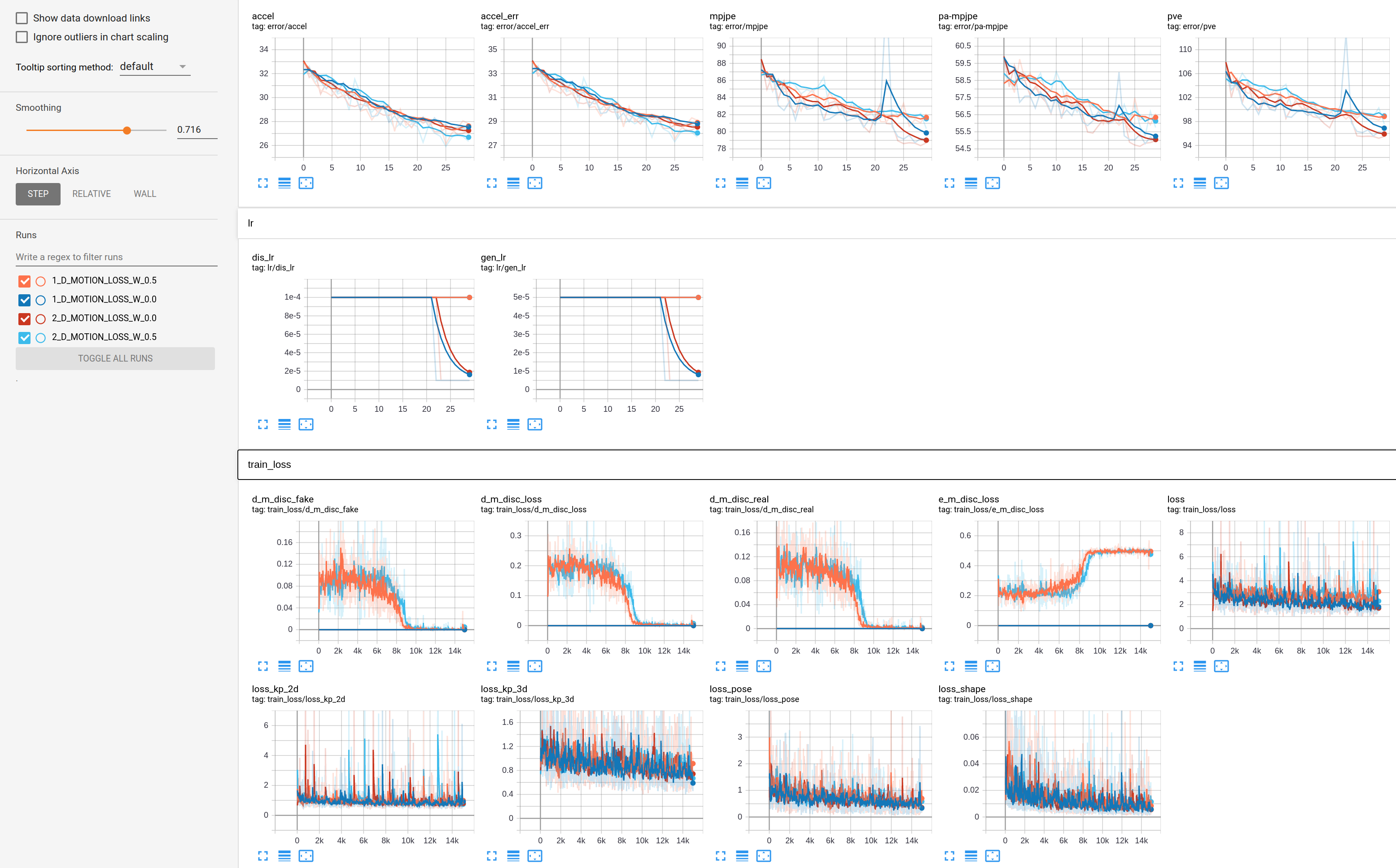
1_D_MOTION_LOSS_W_0.0: train_log.txt 1_D_MOTION_LOSS_W_0.5: train_log_2.txt 2_D_MOTION_LOSS_W_0.0: train_log_3.txt 2_D_MOTION_LOSS_W_0.5: train_log_4.txt
I hope we can resolve these issues soon, and am happy about any suggestions what could be the problem here.
Hi @SDNAFIO,
Thanks for your detailed analysis. Here is my replies some of your comments.
-
The performance of
vibe_model_w_3dpw.pth.taron 3DPW validation set seems indeed weird. Let me inspect this issue and take a closer look what is going on. I will update here. -
Training datasets we are using are quite large and we shuffle them, therefore the results are quite stochastic. It is needed to run multiple experiments with identical seeds for a fair comparison. This is what we did for the motion discriminator experiments.
First of all, I want to ensure the size of evaluation set is 20 or 67? Because in ZephirGe's training log, it's 67.
So maybe there are two kinds of validation set, which lead to different gains on validation performance?
Dear @SDNAFIO and @shyanguan,
After an initial inspection, I noticed that I used some of the validation sequences from 3DPW during data preprocessing by mistake. This was an unintentional mistake I made while copying the data files, I am sorry for that. This explains the ~1-2mm difference when you try to reproduce the results using 3DPW data and the significantly better performance on the 3DPW validation set as @SDNAFIO pointed out.
I am currently running some more experiments. We will do the required corrections for the results affected by this.
Thank you for the quick update! Did this also affect the ablation study in the paper?
Hi @mkocabas @SDNAFIO, is there any update?
Hi @shyanguan,
You can rely on the numbers that you can reproduce for now. We will not do any major changes on the repo since the problem was with the data I used. I am about to finalize the results, after getting those we will make the required updates to the paper and the readme of this repo. Thanks for your patience.
Since almost two months have passed, is there already an update available?
Hi,
Sorry for the late response. I added a note in the evaluation section of the README and I removed the 3DPW training from the code for now.
For the motion discriminator, I ran 25 experiments with and without motion discriminator using the same random seed. The best accuracies obtained are 55.9 (VIBE with MD) vs 56.8 (VIBE without MD) mm PA-MPJPE on 3DPW. An update to ArXiv is on the way.
@mkocabas this is a major progress on VIBE, thank you for the transparency and your open-science approach. A minor technical remark for reproducibility, comparing best accuracy of the experiments is less robust than comparing the median accuracy.
@mkocabas
Could you give us more details about results from Table 2 in the paper?
Did you train using the 3DPW training set in particular for the line 'Baseline (only G)' with SPIN features?
Thanks.

Hi @fabienbaradel,
The final updated version of table 2 is attached here.
So, we removed the results using 3DPW during training. One reason is that; recently all the 3DPW data started to be used for testing with the 3DPW challenge in ECCV 2020. Another reason, it hurts the qualitative results as I noted in the readme.
In this table, all methods use the same training data noted in the experiments section.
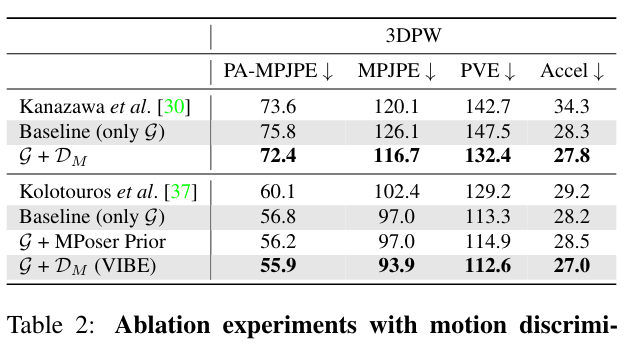
Note that, the results of first 3 row are identical with the previous version since we never used 3DPW training with those methods.
Thanks a lot @mkocabas for your answer :)
@mkocabas Thanks for your answer. But could you give more details on the above table? Exactly which datasets did you use for training the second 3 rows? and what are the bad qualitative results of the method when trained with 3DPW train set?
have someone meet this problems "MPJPE error is 116.87964200973511, higher than 80.0. Exiting!..."? I train this work sometimes meets this.