Include in Docker image necessary drivers for pyodbc
Can you please make it possible to write to Azure SQL Database from this Docker image using pyodbc? It is unable to be installed because of missing drivers. Thanks!
in the conda_deoendcies.yml ; I added something like this: dependencies:
The python interpreter version.
Currently Azure ML Workbench only supports 3.5.2 and later.
- python=3.7.*
Required by azureml-defaults, installed separately through Conda to
get a prebuilt version and not require build tools for the install.
- psutil=5.6 #latest
- pyodbc=4.0.28
Required packages for AzureML execution, history, and data preparation.
- anaconda
- pip ......
and in inference_config.yml u need to add extraDockerfileSteps: (a file include the following:) RUN apt-get -y update && apt-get -y install curl
RUN apt-get -y update &&
apt-get install -y apt-transport-https &&
curl https://packages.microsoft.com/keys/microsoft.asc | apt-key add - &&
curl https://packages.microsoft.com/config/debian/9/prod.list > /etc/apt/sources.list.d/mssql-release.list &&
apt-get update &&
ACCEPT_EULA=Y apt-get install msodbcsql17 unixodbc-dev -y
but I am getting another error now (No module named 'ruamel')
Apologies for the late reply!
Unfortunately, we won't be able to get to this right now. My recommendation is to use your own custom image: https://github.com/microsoft/MLOpsPython/blob/master/docs/custom_container.md
Hi @cindyweng, I was able to resolve this issue by creating a custom "run-config" environment for the ML-Ops Python,
We can install pyodbc and any other pip packages by going to ml_service/pipelines/diabetes_regression_build_train_pipeline.py
On line 37 and 38, we can replace the code with the one I provided below. This will create custom run-config and then manually we can enter the libraries to be used here.
##Defining the run configuration for the Pipeline to run
run_config = RunConfiguration()
# The compute on which the pipeline would be running
# and training the model
run_config.target = aml_compute
run_config.environment.python.conda_dependencies = \
CondaDependencies.create(
conda_packages=['pandas','scikit-learn','numpy','seaborn','matplotlib',
'pyodbc','openpyxl'],pip_packages=['azureml-sdk',
'azureml-dataset-runtime[fuse,pandas]','azureml-defaults',
'azureml-interpret','python-dotenv'],
pin_sdk_version=False)
Code Before:
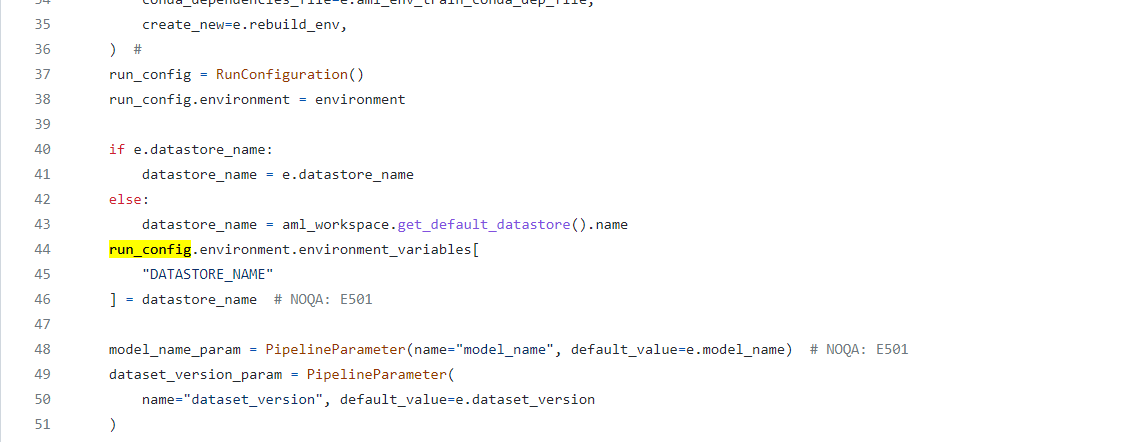
Code After:

I hope this resolves this issue. Contributors: @muditmathur2020 and @OmkarChoulwar
Regards, Mudit Mathur Data Scientist
Hi @cindyweng, I was able to resolve this issue by creating a custom "run-config" environment for the ML-Ops Python,
We can install pyodbc and any other pip packages by going to ml_service/pipelines/diabetes_regression_build_train_pipeline.py
On line 37 and 38, we can replace the code with the one I provided below. This will create custom run-config and then manually we can enter the libraries to be used here.
##Defining the run configuration for the Pipeline to run run_config = RunConfiguration() # The compute on which the pipeline would be running # and training the model run_config.target = aml_compute run_config.environment.python.conda_dependencies = \ CondaDependencies.create( conda_packages=['pandas','scikit-learn','numpy','seaborn','matplotlib', 'pyodbc','openpyxl'],pip_packages=['azureml-sdk', 'azureml-dataset-runtime[fuse,pandas]','azureml-defaults', 'azureml-interpret','python-dotenv'], pin_sdk_version=False)Code Before:
Code After:
I hope this resolves this issue. Contributors: @muditmathur2020 and @OmkarChoulwar
Regards, Mudit Mathur Data Scientist
To summarize, adding any custom library in the environment is possible using the CondaDependencies in the azureml python. sdk.