MACS
 MACS copied to clipboard
MACS copied to clipboard
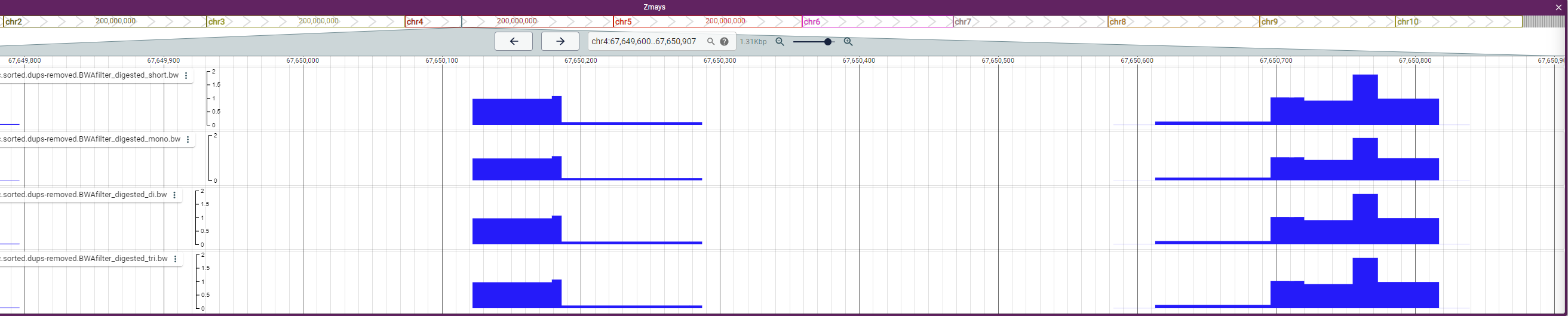
Bug: Same output in all .bdg files when running --save-digested with HMMRATAC
I have tried running macs3 hmmratac. I set the --save-digested flag because I want this for downstream analysis. I note that the bedgraph files contain the same signal in short-, mono-, di-, and tri- signals in all four bedgraph files in all regions.
I have analyzed public ATAC-seq data from https://pubmed.ncbi.nlm.nih.gov/33407087/. Data available at https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP272433&o=acc_s%3Aa samples SRR12261697, SRR12261698, SRR12261699. These all have good periodicity in the insert size distribubion plots so I expect that these would work well.
I have attached a screenshot from a genome browser on what it looks like.
- OS: Linux
- Python version 3.9.12
- Numpy version 1.24.1
- MACS 3.0.0b1
I first tried this on newly generated data from our lab and got the same signal in all four output files.