the batchsize with the gradcache
Dear writer, Your work is very good to me, I want to mix the SimCLR,but I don't know how to do because I find the gradcache without batchsize, but the SimCLR compute the loss function need the batchsize, So I don't how to deal the probelum. please give me some solutions or some tips if you are free.
Thanks advance! Anyway,thanks your work, it solve me a difficulty!
You can define your own loss function and pass it to the GradCache class during initialization. This should give you more fine-grained control over loss computation.
Do note that our standard loss function takes a mean over the batch, as you can see here.
Thank to your reply.
Actually, I have write a loss function,but my loss funciton is different with the SimpleContrastive loss.
My loss funciton equal is NT-Xent loss,similar as follows :
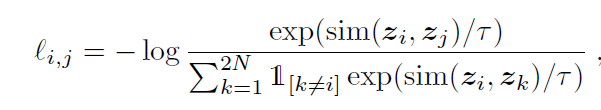
the N is batch size.
the part loss code is ` class NTXentLoss(torch.nn.Module):
def __init__(self, device, batch_size, temperature_or_m, use_cosine_similarity):
super(NTXentLoss, self).__init__()
self.batch_size = batch_size
self.temperature = temperature_or_m
self.device = device
self.softmax = torch.nn.Softmax(dim=-1)
self.mask_samples_from_same_repr = self._get_correlated_mask().type(torch.bool)
self.similarity_function = self._get_similarity_function(use_cosine_similarity)
self.criterion = torch.nn.CrossEntropyLoss(reduction='sum')
def forward(self, zis, zjs):
representations = torch.cat([zjs, zis], dim=0)
similarity_matrix = self.similarity_function(representations, representations)
# filter out the scores from the positive samples
l_pos = torch.diag(similarity_matrix, self.batch_size)
r_pos = torch.diag(similarity_matrix, -self.batch_size)
positives = torch.cat([l_pos, r_pos]).view(2 * self.batch_size, 1)
negatives = similarity_matrix[self.mask_samples_from_same_repr].view(2 * self.batch_size, -1)
logits = torch.cat((positives, negatives), dim=1)
logits /= self.temperature
labels = torch.zeros(2 * self.batch_size).to(self.device).long()
loss = self.criterion(logits, labels)
return loss / (2 * self.batch_size)
`
I know your SimpleContrastiveLoss class,but your loss class seems to have not batch_size.So I am confused how to deal with the batch size in my loss function code. I guess maybe the chunks_size in your code same with the batch_size in my code? If my guess is correct,I just replace batch_size with chunks_size. In a word, I don'w know how to solve the loss function included batch size. Thanks in advance.
By the way,the chunks_size in your code is same with in paaer ? Whether is the chunks_size in your code is the number of subbatch ,or the size of a subbatch?
Oh, I just realized that it is SimCLR that you are talking about. It is a little different from what the example you put here shows. With SimCLR you will have only one encoder and a loss function defined over a single batch of encodings.
As for chunk_size, it refers to sub-batch size in the paper.
Ok,Thanks, I Maybe seems to run it.
yes,the SimCLR only need to a single model.so
gc = GradCache( models=[model,model], # models=[model], # chunk_sizes=8, chunk_sizes=self.config['chunk_size'], loss_fn=self.loss_func, # get_rep_fn=lambda v: v.pooler_output )
I don't whether the gradcache init is true,but it can run.
I noticed that the batchsize can not be set to very large in the case of gradCache, otherwise the batchsize will still be too large. Is that right
If you fail with a large batch, there must be something wrong. (Unless it is million size large, in which case you would probably need to do some off-loading.)
Does the million size refer to the size of the dataset or batch size? If it refer to the size of the dataset, my dataset is one hundred thousand more.
Size of the mini batch for a gradient update.
Very Rarely will this be a problem.