LanczosNetwork
 LanczosNetwork copied to clipboard
LanczosNetwork copied to clipboard
Lanczos Network, Graph Neural Networks, Deep Graph Convolutional Networks, Deep Learning on Graph Structured Data, QM8 Quantum Chemistry Benchmark, ICLR 2019
Lanczos Network
This is the PyTorch implementation of Lanczos Network as described in the following ICLR 2019 paper:
@inproceedings{liao2019lanczos,
title={LanczosNet: Multi-Scale Deep Graph Convolutional Networks},
author={Liao, Renjie and Zhao, Zhizhen and Urtasun, Raquel and Zemel, Richard},
booktitle={ICLR},
year={2019}
}
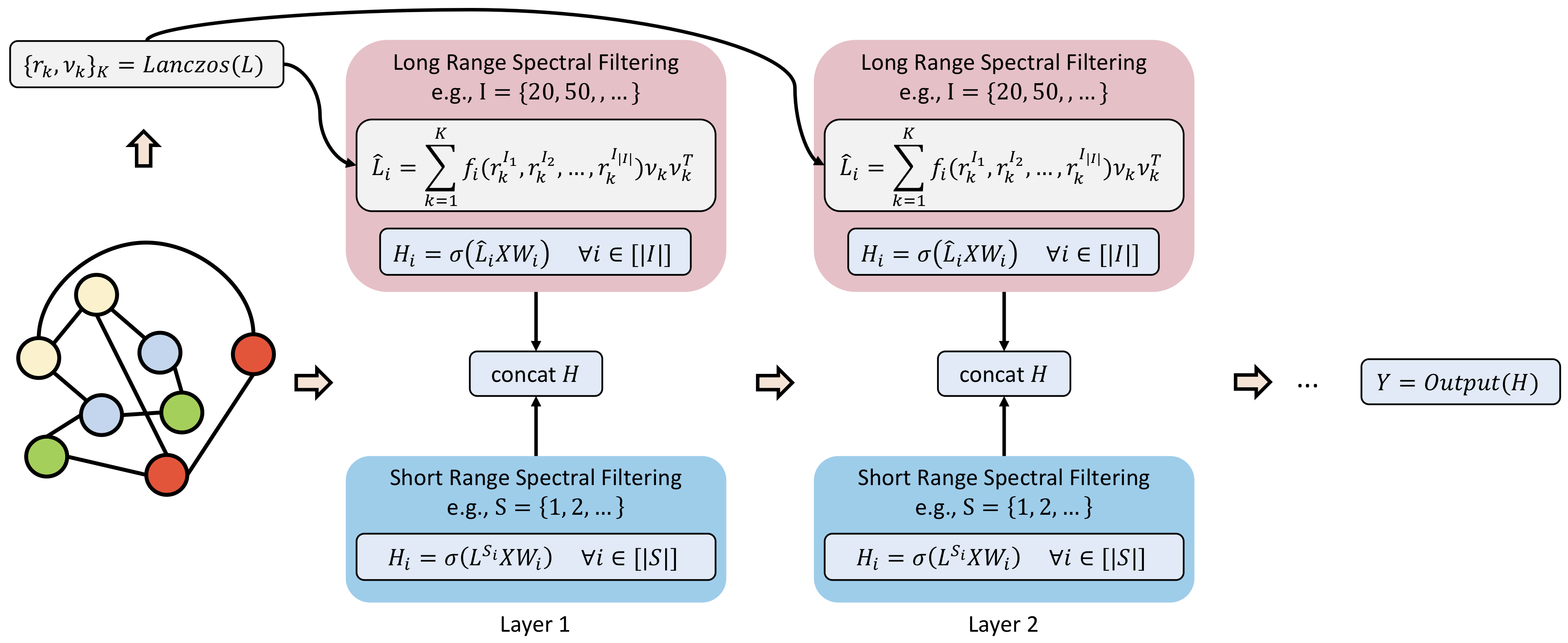
Visualization
Benchmark
We also provide our own implementation of 9 recent graph neural networks on the QM8 benchmark:
- graph convolution networks for fingerprint (GCN-FP)
- gated graph neural networks (GGNN)
- diffusion convolutional neural networks (DCNN)
- Chebyshev networks (ChebyNet)
- graph convolutional networks (GCN)
- message passing neural networks (MPNN)
- graph sample and aggregate (GraphSAGE)
- graph partition neural networks (GPNN)
- graph attention networks (GAT)
You should be able to reproduce the following results of weighted mean absolute error (MAE x 1.0e-3):
| Methods | Validation MAE | Test MAE |
|---|---|---|
| GCN-FP | 15.06 +- 0.04 | 14.80 +- 0.09 |
| GGNN | 12.94 +- 0.05 | 12.67 +- 0.22 |
| DCNN | 10.14 +- 0.05 | 9.97 +- 0.09 |
| ChebyNet | 10.24 +- 0.06 | 10.07 +- 0.09 |
| GCN | 11.68 +- 0.09 | 11.41 +- 0.10 |
| MPNN | 11.16 +- 0.13 | 11.08 +- 0.11 |
| GraphSAGE | 13.19 +- 0.04 | 12.95 +- 0.11 |
| GPNN | 12.81 +- 0.80 | 12.39 +- 0.77 |
| GAT | 11.39 +- 0.09 | 11.02 +- 0.06 |
| LanczosNet | 9.65 +- 0.19 | 9.58 +- 0.14 |
| AdaLanczosNet | 10.10 +- 0.22 | 9.97 +- 0.20 |
Note:
- Above results are averaged over 3 runs with random seeds {1234, 5678, 9012}
Setup
To set up experiments, we need to download the preprocessed QM8 data and build our customized operators by running the following scripts:
./setup.sh
Note:
- We also provide the script
dataset/get_qm8_data.pyto preprocess the raw QM8 data which requires the installation of DeepChem. It will produce a different train/dev/test split than what we used in the paper due to the randomness of DeepChem. Therefore, we suggest using our preprocessed data for a fair comparison.
Dependencies
Python 3, PyTorch(1.0)
Other dependencies can be installed via
pip install -r requirements.txt
Run Demos
Train
-
To run the training of experiment
XwhereXis one of {qm8_lanczos_net,qm8_ada_lanczos_net, ...}:python run_exp.py -c config/X.yaml
Note:
- Please check the folder
configfor a full list of configuration yaml files. - Most hyperparameters in the configuration yaml file are self-explanatory.
Test
-
After training, you can specify the
test_modelfield of the configuration yaml file with the path of your best model snapshot, e.g.,test_model: exp/qm8_lanczos_net/LanczosNet_chemistry_2018-Oct-02-11-55-54_25460/model_snapshot_best.pth -
To run the test of experiments
X:python run_exp.py -c config/X.yaml -t
Run on General Graph Datasets
I provide an example code for a synthetic graph regression problem, i.e., given multiple graphs, each of which is accompanied with node embeddings, it is required to predict a real-valued graph embedding vector per graph.
-
To generate the synthetic dataset:
cd datasetPYTHONPATH=../ python get_graph_data.py -
To run the training:
python run_exp.py -c config/graph_lanczos_net.yaml
Note:
- Please read
dataset/get_graph_data.pyfor more information on how to adapt it to your own graph datasets. - I only add support to LanczosNet by slightly modifying the learnable node embedding to the input node embedding in
model/lanczos_net_general.py. It should be straightforward for you to add support to other models if you are interested.
Cite
Please cite our paper if you use this code in your research work.
Questions/Bugs
Please submit a Github issue or contact [email protected] if you have any questions or find any bugs.