New USGS ESPA Landsat Scene Filenames
When running processLandsatBatch with the new Collection 1 Landsat scene files, the getSceneInfo triggers an error as it does not recognize any of the new file names. The old file names were as such: LT50200461999204-SC20170408040703.tar.gz but the new ones look like this: LT050200461999123001T1-SC20170424204413.tar.gz. I would use an automator to rename the files to get rid of the extra 001T1, but I'm not exactly sure if that's the part that needs to be removed, and the explanation of the filenames on the landsat website (https://landsat.usgs.gov/landsat-collections) isn't exactly accurate so I've emailed customer service as well. Does anyone have suggestions for this?
Also see #64
We're working on that and hope to finalize by next week. The development version of getSceneinfo already works with the new collection and possibly processLandsat too.
You can install the development version of the package with
devtools::install_github('loicdtx/bfastSpatial', ref = 'develop')
The development version isn't working. I recently installed the latest version of R (3.4.0), could this be why? I can't even install the regular version of bfastSpatial either. Are you all working on this as well, or do I need to revert to an older version of R?
I doubt the version of R is the issue. Everything works fine on my system with R 3.4.0. R should tell you where/why the installation fails.
@smithve87 It failed on my computer too, reinstalling rgdal before the installation solved the problem (with force=TRUE option)
I reinstalled everything and all is working now. Also, I was wondering how the update is coming to bfastSpatial to accept the new Landsat collection, is it complete? I tried using the development version, but to no avail.
@smithve87 until the new version is available, below is a "primitive" function to rename the files (gz folders and bands) from Collection 1 standard to Precollection. It works on Mac and should work on Ubuntu
retarVI <- function(VI=c("NDVI"), gzDir=gzDir) {
untarDir <- paste0(gzDir, "/1_untar")
retarDir <- paste0(gzDir, "/2_re_tar")
dir.create(untarDir, showWarnings = FALSE, recursive = TRUE)
dir.create(retarDir, showWarnings = FALSE, recursive = TRUE)
list <- list.files(pattern="*.gz", gzDir, full.names=TRUE)
exDirs <- paste0(untarDir, "/", gsub(".tar.gz", "", substr(basename(list), 1,22)))
#untar function
untar_tiff <- function(filename, exdir) {
dir.create(exdir, showWarnings = F)
tarList <- untar(filename, list=TRUE)
untar(filename, files = str_c(tarList[str_detect(tarList, tolower(VI))]), exdir = exdir)
untar(filename, files = str_c(tarList[str_detect(tarList, "MTL")]), exdir = exdir)
untar(filename, files = str_c(tarList[str_detect(tarList, "cfmask_conf")]), exdir = exdir)
untar(filename, files = str_c(tarList[str_detect(tarList, "cfmask")]), exdir = exdir)
}
#rename function
rename.my.files <- function(from, to) {
todir <- dirname(to)
if (!isTRUE(file.info(todir)$isdir)) dir.create(todir, recursive=TRUE)
file.rename(from = from, to = to)
}
for (i in 1:length(list)) {
#1. untar
untar_tiff(filename = list[i], exdir = exDirs[i])
#2. renaming folder
MTLlist <- list.files(pattern="*MTL.txt", exDirs[i], full.names=TRUE)
x <- read.table(MTLlist, quote="\"", comment.char="", stringsAsFactors=FALSE, sep = "=", nrows=5)
preCollectionsceneID <- str_trim(x[5,2])
C1SceneID <- gsub("_MTL.txt", "", basename(MTLlist))
oldFolder <- exDirs[i]
newFolder <- paste0(dirname(exDirs[i]), "/", preCollectionsceneID)
rename.my.files(from=oldFolder, to=newFolder )
#renaming bands
oldBands <- list.files(path=newFolder, pattern="*.tif", full.names = TRUE)
newBands <- gsub(C1SceneID,preCollectionsceneID,oldBands)
for (j in 1:length(oldBands)){
rename.my.files(from=oldBands[j], to=newBands[j])
}
#3. re-tar
setwd(newFolder)
retarfile <- paste0(retarDir, "/", basename(newFolder), ".tar")
gzfile <- paste0(retarDir, "/", basename(newFolder), ".tar.gz")
pathToFiles <- paste0(untarDir, "/", basename(newFolder))
command <- paste("tar -cvzf " , gzfile, " -C ", pathToFiles, " . " )
system(command=command, intern = FALSE,ignore.stdout = TRUE, ignore.stderr = FALSE)
#delete intermediar files
setwd(gzDir)
unlink(newFolder, recursive = TRUE)
}
}
Thanks @mabdelrazek, I'll give it a shot this weekend if I can (R is bogged down running other things), and let you know how it works.
I'm on it right now, hang tight, there will be a pre-release very soon
@loicdtx Thanks for the effort!
I just updated the development version of the package. It includes a new version of processLandsat that should work for both Landsat collections. It's not fully tested yet, but I did a few runs with different configurations and it seems to work well.
Take a look at the updated doc too cause a few things have changed. At least:
vican now receive an vector of 'indices' (e.g.:vi = c('ndvi', 'evi', 'tcb', 'sr_band3'))srdircan be omitted (in which case the bands used for computing the indices are temporarily stored in a tmp directory)- No more
untar
To install the development version of the package:
devtools::install_github('loicdtx/bfastSpatial', ref = 'develop')
Example usage:
library(bfastSpatial)
processLandsat(x = '/path/to/landsat/archive.tar.gz',
outdir = '/home/user/sandbox/landsat_processing',
vi = c('ndvi', 'tcg'), mask = 'cfmask', keep = c(0),
delete = TRUE, overwrite = TRUE)
Let us know if you encounter problems or if you have any suggestions.
Thanks @loicdtx ! You saved us a huge time by doing this 👍 I also find the possibility to process several VIs at in one line of code quite nice. You should definitely add the package to CRAN
I can't get this version to work...
I tried the "processLandsat" call from @loicdtx above, but no dice.
I get an error of: "Error in if (x == "" | x == ".") { : argument is of length zero"
Apparently this happens if a NULL value is compared in an IF statement, but I can't tell where.
It seems to get through the .createDirs function call, because I can see the directories created, but I don't exactly know how far it gets.
If anyone can help, I'd appreciate!
Thanks!
@mtnorton are you working with espa data? Does your data archive include the cfmask band?
@loicdtx Actually there is no 'cfmask' - we downloaded the data after the change on June 2nd.
I guess I can hack through the data processing myself if that's all it is, and use the bfm function on that.
@mtnorton Yes, or use processLandsat(Batch) with the pixel_qa layer as mask.
# Landsat 5-7
processLandsat(x = '/path/to/landsat/archive.tar.gz', outdir = tmpDir(),
vi = 'ndvi', delete = TRUE, mask = 'pixel_qa', keep = c(66, 130),
overwrite = TRUE)
# Landsat 8
processLandsat(x = '/path/to/landsat/archive.tar.gz', outdir = tmpDir(),
vi = 'ndvi', delete = TRUE, mask = 'pixel_qa', keep = c(322, 386),
overwrite = TRUE)
Thanks @loicdtx, got it working. I pulled out NDVI from the tarballs using that code above and everything seems to be working correctly.
The only thing I'd note is that the saturated pixels (value=20000) aren't filtered by default. It's a problem because they trigger breaks within the BFAST algorithm, even for a single observation.
Yes, fill value filtering is on my todo list (see #72).
Once you have created your RasterBrick using TimeStack, you can filter these values using the code below:
cleanFun <- function(x) {
x[x == 20000] <- NA
return(x)
}
infile <- 'data/ndvi_stack.grd'
outfile <- ''data/ndvi_stack_clean.grd''
z <- getZ(brick(infile))
ndvi_stack <- brick(infile) %>%
calc(fun = cleanFun, filename=outfile, datatype='INT2S')
ndvi_stack <- setZ(ndvi_stack, z)
hdr(ndvi_stack, 'RASTER')
@loicdtx, @mtnorton and @mabdelrazek - Thanks for all of your help and insight thus far. I haven't gotten around to messing with cleaning the fill/saturation values, but the dev version of bfmSpatial seems to be working. However, I've run into a couple issues I think. I've gone through everything step by step to try and pinpoint errors, and found some hiccups with the sensor / s subsetting, as well as with the resulting raster stack produced by timeStack.
With the sensor, I messed with having sensor contain c("ETM+ SLC-on", "ETM+ SLC-off"), which is the default, as well as c("ETM+ SLC-on", "ETM+ SLC-off", "OLI") for the OLI Landsat 8 images. They produce slightly difference results, so I'm wondering what exactly the difference is, because even when OLI is not included it seems like the Landsat 8 images are included. If someone could shed some light on this I'd appreciate it.
The timeStack issue is a bit stranger. When I run timeStack on a directory that contains both new and old Landsat file formats, it creates a brick with the proper number of layers, but the dates are off:
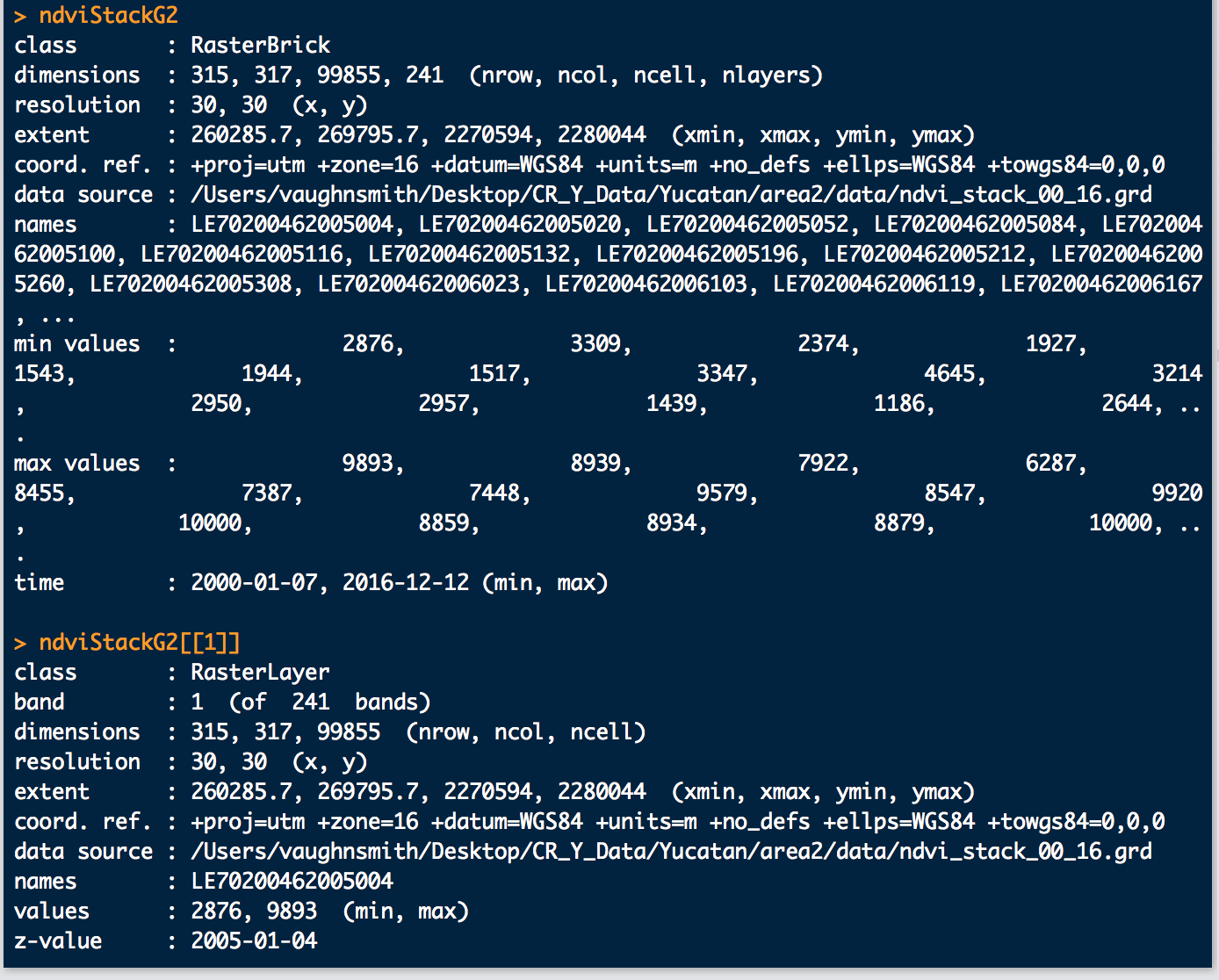
Also, these are how the dates look:
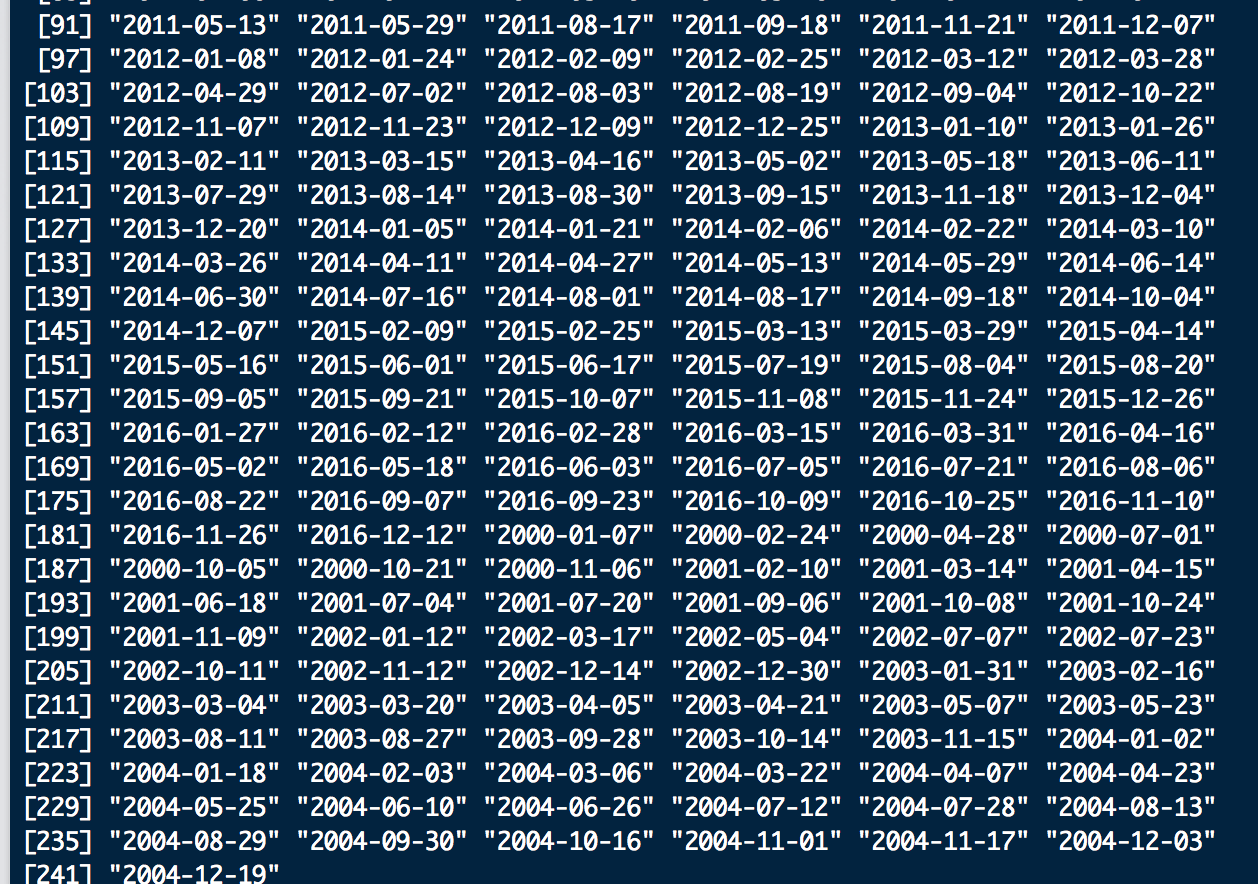
As you can see the brick shows the correct time 2000 - 2016, but the first layer of the brick is 2005 so this will throw of the model since it starts in the middle and the actual beginning of the time series is at the end. The 2000-2004 data is the new Landsat file format data, so it seems to be throwing off timeStack somehow. Will this matter in the end, or does bfmSpatial / bfastmonitor use the min/max date, or just automatically order by date as opposed to by brick layer? It's difficult to tell from resulting outputs, but I'm digging into the code now to see what I can find. If someone could again shed some light on this I would very much appreciate it.
Thank you!
@smithve87 I noticed the same thing, but it seemed as if the results were coming out in the proper order. I dove in to see where the "order" function call was, but couldn't find it. It does seem a little weird in terms of the getZ results being out of order, but I think everything worked out correctly for me.
That's correct; the layer order does not mater for functions like bfmSpatial, as long as the time of each layer is properly set. This is a bug with orderChronoFun (a sub-function of timeStack) that should have been edited to support the new collections but didn't.
@smithve87 Which function(s) are you referring to with:
With the sensor, I messed with having sensor contain c("ETM+ SLC-on", "ETM+ SLC-off"), which is the default, as well as c("ETM+ SLC-on", "ETM+ SLC-off", "OLI") for the OLI Landsat 8 images. They produce slightly difference results, so I'm wondering what exactly the difference is, because even when OLI is not included it seems like the Landsat 8 images are included. If someone could shed some light on this I'd appreciate it.
@mtnorton & @loicdtx Good to know about the order of the layers so thanks for letting me know. As for the sensor, I'm referring to that optional reformatting call:
optional: reformat sensor if needed if("ETM+" %in% sensor) sensor <- c(sensor, "ETM+ SLC-on", "ETM+ SLC-off")
As well as the beginning of:
fun <- function(x) { # subset x by sensor if(!is.null(sensor)) x <- x[which(s$sensor %in% sensor)] ...
So the raster brick x gets subset by sensor, and subsetting seems to eliminate some spurious breaks. I've run bfmSpatial and subset by sensor with and without the OLI sensor for LS8 images, and get varied results. It seems like the LS8 / OLI sensor images / layers are included, but I've had runs where I didn't include OLI within sensor and the 2016 data didn't show up in the results. I guess I'm just curious about how subsetting works in this situation.