umap crashes in my computer with 900,000 points
Hi, I have been trying to embed 900,000 points using UMAP in my computer. The program eventually gets killed by the system. I tried running in both Jupyter and in terminal.
My system: 16Core/32Thread AMD CPU, 128GB RAM (Terminal reports 125GB). Ubuntu 18.04.3 LTS.
I was wondering if it is a system requirement issue or an issue in how the UMAP handles this many points. (In the paper, it seems UMAP can handle millions of points as there is a visualization of 3Million points.)
Here is a code that reproduces the error in my computer:
import numpy as np
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
X_main = np.random.rand(900000, 1000)
n_components = 2
pca = PCA(n_components = 50)
X_train = pca.fit_transform(X_main)
n_neighbors= 50
MIN_DIST = 0.1
import umap
ump = umap.UMAP(n_neighbors=n_neighbors,
min_dist=MIN_DIST,
n_components=2,
random_state=100,
metric= 'euclidean')
y_umap = ump.fit_transform(X_train)
The most likely reason for a silent crash with the system killing the job is a memory issue. UMAP can be pretty memory hungry (newer development versions are working to fix this). At least one option is the try the option low_memory=True which will try to use a sometimes slower but less memory hungry approach. Another option is to install the latest (version 0.5 or newer) version of pynndescent.
Thanks. I will try pynndescent.
I also think it overflows memory. I tried to compute an accurate nearest neighbor matrix for this data and observed crashing.
The following overflows memory:
@numba.jit(nopython=True, parallel=True)
def nn_neib_1(X,n_neighbor=10):
N = X.shape[0]
N_neib_array = np.zeros((N,n_neighbor), dtype=np.int32)
for i in prange(N):
dist = np.sum((X - X[i])**2,axis=1)
N_neib_array[i] = np.sort(np.argsort(dist)[1:n_neighbor+1])
return N_neib_array
But this one works just fine:
@numba.jit(nopython=True, parallel=True)
def nn_neib_2(X,n_neighbor=15):
N = X.shape[0]
N_neib_array = np.zeros((N,n_neighbor), dtype=np.int32)
for i in range(N):
if (i+1)%10000 == 0:
print('Completed ', i+1, ' of ', N)
dist = np.zeros((N), dtype=np.float32)
for j in prange(N):
dist[j] = np.sum( (X[i]-X[j])**2 )
dist[i] = -1
N_neib_array[i] = np.sort(np.argsort(dist)[1:n_neighbor+1])
return N_neib_array
So I guess piecewise parallelizing the process will help a lot.
Hello :)
I have a similar although more mysterious issue. I am trying to obtain 2D embeddings of various representations generated by the layers of of a Recurrent Neural Network (LSTM).
UMAP gracefully embed the representation generate by one of the layers, a matrix of size 250k X 120, but silently crash with the representation generated by another layer, a smaller 250k X 80 matrix.
Precisely, my python program is killed at this stage:
UMAP(angular_rp_forest=True, low_memory=False, metric='cosine', min_dist=0.8, n_epochs=1000, n_neighbors=100, verbose=True)
Mon Sep 20 09:30:15 2021 Construct fuzzy simplicial set
Mon Sep 20 09:30:16 2021 Finding Nearest Neighbors
Mon Sep 20 09:30:16 2021 Building RP forest with 30 trees
What I've done so far:
- Checked for NaNs.
- Checked for extremely large or small numbers.
- Reduced the
n_neighborsparameters to 50 - Tried with PCA (which runs successfully), here an image of the PCA reduction (colors are different "categories" and it makes sense for them to have such an evident separation).
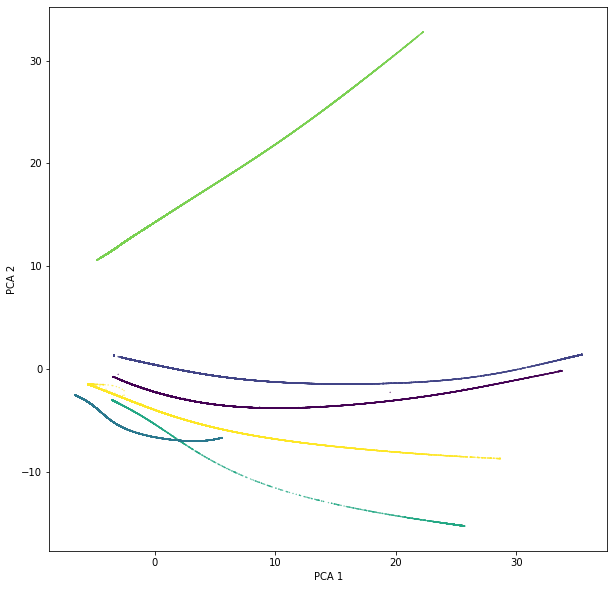
I am running this on:
- Windows OS machine
- Python 3.6.8
- UMAP master branch on GitHub from 1 month ago
- pynndescet 0.5.2
It may just be a memory issue -- as in not enough of it. UMAP can be pretty memory hungry when doing nearest neighbor computations, and depending on that dataset that can get very expensive. I would definitely try with low_memory=True as that will likely help a little.
Thank you very much for the swift response! I should have expanded a bit:
- I tried with
low_memory=Trueas well but with no success. - I assume at this point that the amount of required memory does not only depend on the size of the original dataset but also on its "characteristics" since I was able to embed the
250k X 120matrix (108MB) but not the250k X 80one (84MB).
Apologies for the silly questions but I am trying to understand how to handle this type of situations.
I'm not sure if this is correct, if you are running two UMAP instances one after another, some memory from previous instances may still be occupied. But chance of this happening may be low.
Also, you may check whether the issue is happening in UMAP or pynndescent. pynndescent is sometimes dataset dependent. I vaguely remember a twitter thread regarding this.
Thank you for your answers! I have a couple of new insights on this:
- Running multiple consecutive instances of UMAP doesn't seem to be the problem (I can easily reduce many large datasets one after the other without running into memory problems).
- Changing the distance metric from "cosine" to "euclidean" did solve the silent crash problem (I assume because is less expensive?).