PureCN cell line output from Sanger DepMap
Hi Markus,
Could you help me understanding the PureCN output generated by Sanger's DepMap for cell lines? The output can be found here. I picked the representative records that I am confused with (data of SIDM00673 model_id corresponding to HCC70; many other segment outputs does make sense to me). Likely this is the callLOH output.
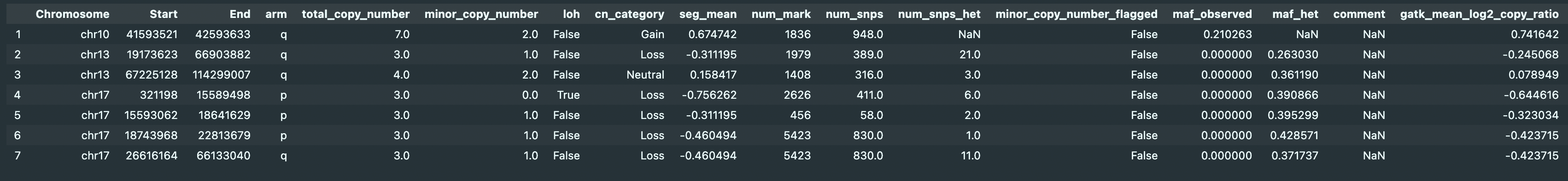
My questions are:
- How "maf_observed" can be 0? I thought as long as "num_snps_het" > 0, there will be non zero AF for it. (I could be completely misunderstood).
- The 6th, and 7th rows seems to be LOH to me. But it actually detected 1 copy of minor allele as compared to 4th row (LOH). On the other hand, 2nd and 5th rows were deem 1 copy for minor allele. Is it because
num_snps_het / num_snpsis relatively high? - How does pureCN found 1st and 4th rows to have 2 copy for minor allele?
There are very few information on how they processed the calls. I sincerely appreciate your comments.
Thank you very much, Hanbin
Apologies @hanbinlu for the way too long delay. I will look into that example in the coming days. It's been a while I worked with cell lines.
Yes, this is indeed a bug. The maf expected and observed are wrong with --model-homozygous.
Should be fixed in current developer branch. Will backport to stable in a week or two. Please let me know if you find other issues with cell line data.