extractSequence function doesn't work
Hi,
I met one problem with the extractSequence function.
My transcript-level counts were produced from salmon. According to the user guide
If you are “only” analyzing known isoforms from a trusted annotation database (if you have used Kallisto/Salmon and not used StringTie etc) you can most likely skip this step as the importRdata() function automatically imports CDS regions from the GTF supplied.
I can run extractSequence directly since I have ORF information in my Switchlist object. I double checked if ther orfanalysis is in my Switchlist object or not. The answer is absolutely yes.
However, when I ran extractSequence function, one error message came out:
Error in extractSequence(dtu_result, writeToFile = T, removeLongAAseq = T, : The genomeObject argument must be a BSgenome
It seems I have to clarify the genomeObject argument. Then, I specified this argument and ran the function again. Another error message appeared.
Step 1 of 3 : Extracting transcript nucleotide sequences... Error in loadFUN(x, seqname, ranges) : trying to load regions beyond the boundaries of non-circular sequence "chr17"
I couldn't debug this error but I found there are lots of NA values in my orfanalysis list. I suspect this may be the possible reason of why extractSequence function doesn't work.
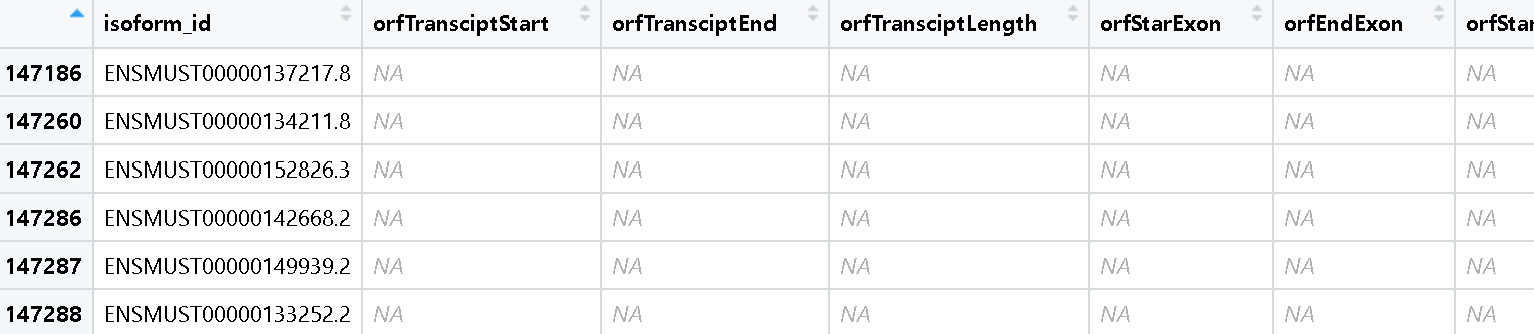
Could you help me figure out this problem?
Thanks, Feng
This looks like the error you would get if you use a wrong genome or genome version. Could you double-check that?
And it is not an error that there are NA values in the ORF annotation (as long as the fraction of them is not to large)?
Also if you have the transcript nucleotide fasta corresponding to the transcripts quantified I'd adding that in the importRdata() step.
Closing due to no activity. If you respond I will open it again :)