keras-unet
 keras-unet copied to clipboard
keras-unet copied to clipboard
Helper package with multiple U-Net implementations in Keras as well as useful utility tools helpful when working with image semantic segmentation tasks. This library and underlying tools come from mul...
About
Helper package with multiple U-Net implementations in Keras as well as useful utility tools helpful when working with image segmentation tasks
Features:
- [x] U-Net models implemented in Keras
- [x] Vanilla U-Net implementation based on the original paper
- [x] Customizable U-Net
- [x] U-Net optimized for satellite images based on DeepSense.AI Kaggle competition entry
- [x] Utility functions:
- [x] Plotting images and masks with overlay
- [x] Plotting images masks and predictions with overlay (prediction on top of original image)
- [x] Plotting training history for metrics and losses
- [x] Cropping smaller patches out of bigger image (e.g. satellite imagery) using sliding window technique (also with overlap if needed)
- [x] Plotting smaller patches to visualize the cropped big image
- [x] Reconstructing smaller patches back to a big image
- [x] Data augmentation helper function
- [x] Notebooks (examples):
- [x] Training custom U-Net for whale tails segmentation
- [ ] Semantic segmentation for satellite images
- [x] Semantic segmentation for medical images ISBI challenge 2015
Installation:
pip install git+https://github.com/karolzak/keras-unet
or
pip install keras-unet
Usage examples:
- U-Net implementations in Keras:
- Vanilla U-Net
- Customizable U-Net
- U-Net for satellite images
- Utils:
- Plot training history
- Plot images and segmentation masks
- Get smaller patches/crops from bigger image
- Plot small patches into single big image
-
Reconstruct a bigger image from smaller patches/crops
Vanilla U-Net
Model scheme can be viewed here
from keras_unet.models import vanilla_unet
model = vanilla_unet(input_shape=(512, 512, 3))
[back to usage examples]
Customizable U-Net
Model scheme can be viewed here
from keras_unet.models import custom_unet
model = custom_unet(
input_shape=(512, 512, 3),
use_batch_norm=False,
num_classes=1,
filters=64,
dropout=0.2,
output_activation='sigmoid')
[back to usage examples]
U-Net for satellite images
Model scheme can be viewed here
from keras_unet.models import satellite_unet
model = satellite_unet(input_shape=(512, 512, 3))
[back to usage examples]
Plot training history
history = model.fit_generator(...)
from keras_unet.utils import plot_segm_history
plot_segm_history(
history, # required - keras training history object
metrics=['iou', 'val_iou'], # optional - metrics names to plot
losses=['loss', 'val_loss']) # optional - loss names to plot
Output:
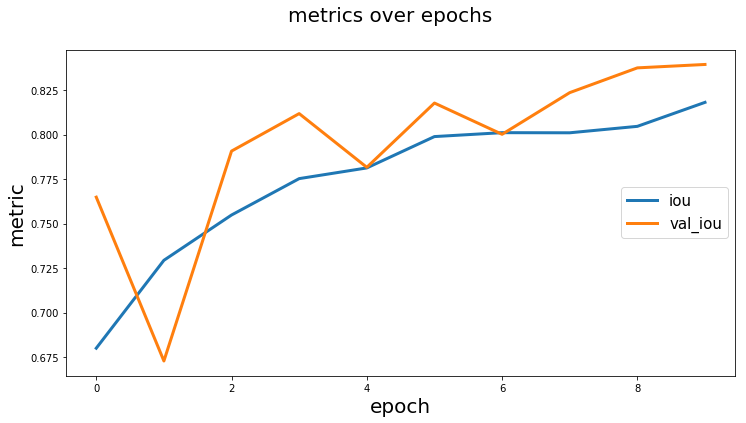
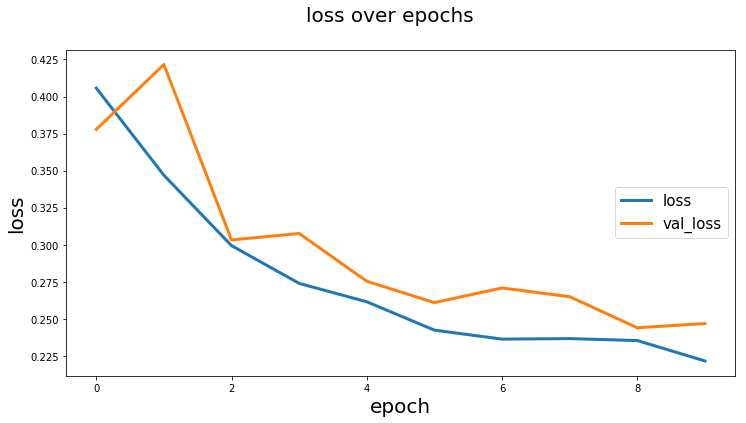
[back to usage examples]
Plot images and segmentation masks
from keras_unet.utils import plot_imgs
plot_imgs(
org_imgs=x_val, # required - original images
mask_imgs=y_val, # required - ground truth masks
pred_imgs=y_pred, # optional - predicted masks
nm_img_to_plot=9) # optional - number of images to plot
Output:
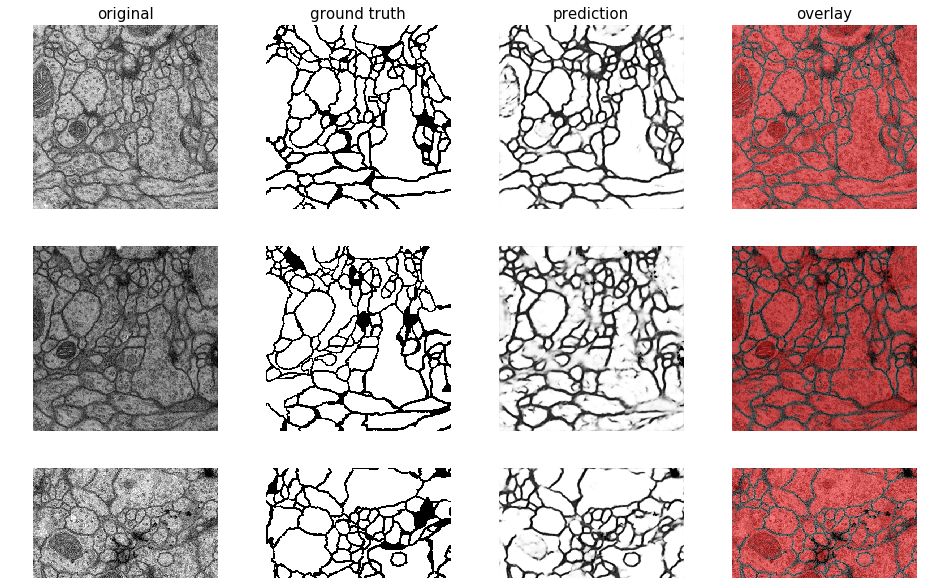
[back to usage examples]
Get smaller patches/crops from bigger image
from PIL import Image
import numpy as np
from keras_unet.utils import get_patches
x = np.array(Image.open("../docs/sat_image_1.jpg"))
print("x shape: ", str(x.shape))
x_crops = get_patches(
img_arr=x, # required - array of images to be cropped
size=100, # default is 256
stride=100) # default is 256
print("x_crops shape: ", str(x_crops.shape))
Output:
x shape: (1000, 1000, 3)
x_crops shape: (100, 100, 100, 3)
[back to usage examples]
Plot small patches into single big image
from keras_unet.utils import plot_patches
print("x_crops shape: ", str(x_crops.shape))
plot_patches(
img_arr=x_crops, # required - array of cropped out images
org_img_size=(1000, 1000), # required - original size of the image
stride=100) # use only if stride is different from patch size
Output:
x_crops shape: (100, 100, 100, 3)

[back to usage examples]
Reconstruct a bigger image from smaller patches/crops
import matplotlib.pyplot as plt
from keras_unet.utils import reconstruct_from_patches
print("x_crops shape: ", str(x_crops.shape))
x_reconstructed = reconstruct_from_patches(
img_arr=x_crops, # required - array of cropped out images
org_img_size=(1000, 1000), # required - original size of the image
stride=100) # use only if stride is different from patch size
print("x_reconstructed shape: ", str(x_reconstructed.shape))
plt.figure(figsize=(10,10))
plt.imshow(x_reconstructed[0])
plt.show()
Output:
x_crops shape: (100, 100, 100, 3)
x_reconstructed shape: (1, 1000, 1000, 3)

[back to usage examples]



