nbconvert
 nbconvert copied to clipboard
nbconvert copied to clipboard
Widgets not displaying properly in HTML
I am trying to get widget.output() to display a table so that I can use HBox and VBox on the widget. Currently this code actually does display 2 tables side by side in the notebook:
import pandas as pd
from IPython.display import display
from ipywidgets import HBox, VBox
import ipywidgets as widgets
f = widgets.Output()
with f:
display(pd.read_csv('Book1.csv'))
display(HBox([f,f]))

However, the problem arises when I try to nbconvert the notebook to html which gives me this

Anyone know what the issue here might be?
The command I am using to convert is: jupyter nbconvert --execute file.ipynb
Just to add my voice to this - I have the same issue: anything capture in Output - which displays in the correct locations in jupyter notebook & lab - is actually displayed immediately under the currently active cell when executing with nbconvert.
I'll have to look into what the Output widget is doing. It's possibly it's relying on there being a javascript engine, or is reusing display ids but I'll need to spend a little time diagnosing to know for sure.
that would be super helpful, thanks. Let me know if I can help.
Sorry for the delay in getting to this.
So the root problem is that nbconvert never had the ability to parse application/vnd.jupyter.widget-view+json message types, which Output() use for display purposes. Only the widget javascript appears to have that encoded anywhere.
From reading the messages sent from the kernel it appears the widgets are using the comm_id content type to capture outputs associated with a widget output (and skipping rendering to the cell), then using a method -> update call setting the msg_id to an empty string to indicate capture is done.
'content': {'comm_id': 'be6d5eaa0c0345c9bb8cb7c902bc451b',
'data': {'buffer_paths': [],
'method': 'update',
'state': {'msg_id': '9412f859-595a00f31a798b3423c847ec'}}},
then
{'buffers': [],
'content': {'data': {'text/html': ...
then
{'buffers': [],
'content': {'comm_id': 'be6d5eaa0c0345c9bb8cb7c902bc451b',
'data': {'buffer_paths': [],
'method': 'update',
'state': {'msg_id': ''}}},
saving the wdiget outputs so that
content': {'data': {'application/vnd.jupyter.widget-view+json': {'model_id': 'be6d5eaa0c0345c9bb8cb7c902bc451b',
'version_major': 2,
'version_minor': 0},
'text/plain': 'Output()'},
will re-render said output.
@jasongrout Can you help confirm expected behavior here? I think I see the pattern being used but I'm not aware if it's documented anywhere and want to make sure I've got it right before improving the comm message processing here.
@MSeal - you correctly figured out how output widgets work - they set the msg_id state attribute to the parent message id that should be captured in the frontend, then later clear the msg_id attribute to indicate capture should end. The frontend output widget should capture any output messages during that time having that specific msg_id as the parent_header.msg_id. The frontend output widget should set its state dictionary to the contents of the relevant output messages (which will then be reflected back to the kernel as part of the state of the widget).
It is the frontend's responsibility to provide an output widget that knows how to interface with the other parts of the frontend to capture relevant outputs. How does nbconvert typically deal with widgets?
It is the frontend's responsibility to provide an output widget that knows how to interface with the other parts of the frontend to capture relevant outputs. How does nbconvert typically deal with widgets?
Messily. Today it mostly captures the com messages, saving the content.data.state and content.data.buffer_paths in the metadata of the notebook so when a UI widget loads it has some state to load from. For conversion to other formats it relies on the widget backends to emit some renderable output (even if a default / non-interactive variant) alongside the backing widget state. This means each new widget, and some of the unmaintained ones, usually don't render anything useful for headless execution.
I noticed in this case the Output widget doesn't save any state to the notebook to resaturate the widget on page reload. It leaves a widget view media typed object in the notebook but no data for what populates said view, which means running from a UI (or nbconvert) doesn't persist what the user sees and instead prints this wherever a widget was:
A Jupyter widget could not be displayed because the widget state could not be found. This could happen if the kernel storing the widget is no longer available, or if the widget state was not saved in the notebook. You may be able to create the widget by running the appropriate cells.
In the case above outlined by the issue, it's accidentally printing one of the tables because nbconvert doesn't capture the initial rendering to be used as the view. I think I could correctly capture it and apply the view where-ever it's referenced at the end, but I'm not sure if there's additional structured patterns that depend on the widget for how to arrange or modify those outputs.
I think it might work for the nbconvert output widget to just capture the output messages sent and put those in the output area's state, similar to how the classic notebook output widget and the jlab output widget behave. Essentially, you'd need to mock this frontend behavior for the nbconvert output widget.
Alternatively, you can use the output widget if you explicitly append to its state from the kernel. That does not rely on capturing from the frontend - see the example in the docs:
out = widgets.Output(layout={'border': '1px solid black'})
out.append_stdout('Output appended with append_stdout')
out.append_display_data(YouTubeVideo('eWzY2nGfkXk'))
out
Here is some more context about the output widget and its design decisions: https://github.com/jupyter-widgets/ipywidgets/issues/2377
We had the same problem in voila: https://github.com/voila-dashboards/voila/pull/91/files
Which resulted in a server side Output widget: https://github.com/voila-dashboards/voila/blob/8557d9d270100e55a40911e1afe61046736eba18/voila/execute.py#L41
I'm not sure this code should live in nbconvert instead, but with some traitlets-foo you may be able to configure nbconvert to use the VoilaExecutePreprocessor to execute your notebook.
Ok I'll move this issue to nbclient and see about taking a stab at mimicing the front-end behavior if possible. Thanks for the context.
This is now solved in nbclient, which means nbconvert 6.0 alpha has the fix.
@MSeal I was having the same issue as the original poster but using Altair plots instead of pandas Dataframe. After checking this issue I upgraded nbconvert from 5.6.1 to 6.0.0a3 using:
pip install nbconvert==6.0.0a3
After the upgrade I tested the original poster's example and it worked as expected.
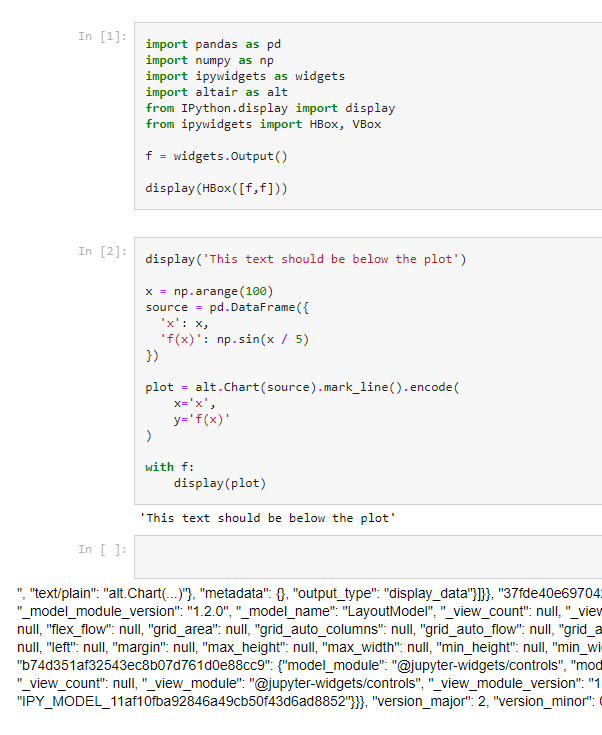
But now, after upgrading, not only the plot is not displayed in the correct place, but it's not displayed at all.
Here is a simple working code that can be used to see the issue (in the actual project we use ipywidgets tabs, and many other widgets where we want to load the plots) as well as how the HTML file looked in Chrome before and after upgrading from 5.6.1 to 6.0.0a3:
import pandas as pd
import numpy as np
import ipywidgets as widgets
import altair as alt
from IPython.display import display
from ipywidgets import HBox, VBox
f = widgets.Output()
display(HBox([f,f]))
display('This text should be below plot')
x = np.arange(100)
source = pd.DataFrame({
'x': x,
'f(x)': np.sin(x / 5)
})
plot = alt.Chart(source).mark_line().encode(
x='x',
y='f(x)'
)
with f:
display(plot)
5.6.1

6.0.0a3
Additional info
Versions:
[5.6.1]
> jupyter --version
jupyter core : 4.6.1
jupyter-notebook : 6.0.3
qtconsole : 4.7.4
ipython : 7.12.0
ipykernel : 5.1.4
jupyter client : 6.1.3
jupyter lab : not installed
nbconvert : 5.6.1
ipywidgets : 7.5.1
nbformat : 5.0.4
traitlets : 4.3.3
[6.0.0a3]
> jupyter --version
jupyter core : 4.6.1
jupyter-notebook : 6.0.3
qtconsole : 4.7.4
ipython : 7.12.0
ipykernel : 5.1.4
jupyter client : 6.1.3
jupyter lab : not installed
nbconvert : 6.0.0a3
ipywidgets : 7.5.1
nbformat : 5.0.4
traitlets : 4.3.3
The following commands were used to execute and convert the notebook to html:
jupyter nbconvert test_nbclient_fix.ipynb --to ipynb --execute --output test_fix.ipynb
jupyter nbconvert test_fix.ipynb --to html --output-dir /path/to/output_dir/ --output test_fix.html
Thanks for reporting and providing a reproducible error. I noticed what's happening is that the child displays are present notebook metadata->widgets namespace, but the children subkey has IPY_MODEL_c72337cc98a94de5adcf9bed7653fb7b which isn't resolving to the c72337cc98a94de5adcf9bed7653fb7b widget chart id correctly when we generate the cell output. I'll need to dig in later to figure out why.
@maartenbreddels may be interested in this as he was helping with the translation of logic from viola
Hey @MSeal @maartenbreddels, I just wanted to check if there has been any updates regarding this. Otherwise maybe you could throw some hints on a temporal workaround? Or it's not possible?
Thank you for the efforts and great work!
Yes, this issue was resolved in https://github.com/jupyter/nbclient/issues/24 and nbclient >= 0.4.0 has the fix. NBconvert 6.0 (which should be releasing tomorrow) defaults to using nbclient and the issue overall should disappear. You can try it out on the 6.0.0rc0 release today if you like.
Hey @MSeal, I tried again with my reproducible error in my preveious response and nothing changed. I still get the same error with 6.0.0rc0 as I did with 6.0.0a3. Maybe you were refering to the original issue that was already solved but not mine?
I am getting the same error as @rvEnertis using 6.0.0rc0 and nbclient 0.5.0 with altair plots
Ok I'll need to dig into why that's happening for altair outputs specifically, might take me a couple days to find time to dig into it though as my free time is pretty limited.
Hi guys, any updates on this? @MSeal @maartenbreddels
I still have the problem of not visible Output widgets content in converted HTML, nbconver version 6.0.7. I use JupyterLab. Does anyone have a workaround?
Hello, I have the same issue while using matplot lab. The widget displays in ipython notebook, but while rendering with nbconvert into html, i dont see them there.
from IPython.display import HTML
import ipywidgets as widgets
%matplotlib inline
from IPython.display import display
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
import warnings
warnings.filterwarnings('ignore')
# plot known markers in tab
celltypes = list(marker_genes.keys())
sub_tab=[widgets.Output() for i in range(len(celltypes))]
tab = widgets.Tab(sub_tab)
rcParams['figure.figsize'] = 3, 3
for i in range (len(celltypes)):
celltype = celltypes[i]
tot = len(marker_genes[celltype])
cols = 4
rows = tot // cols
rows += tot % cols
tab.set_title(i, celltype)
with sub_tab[i]:
print('\033[1m' + celltype)
sc.pl.umap(adata_pp, color = marker_genes[celltype], cmap='icefire', ncols=6, size=1,
legend_fontsize = 8, legend_fontweight = 1)
display(tab)
It is displays in notebook.
 While using
While using
os.system('jupyter nbconvert --to html FindingMarker_1.4.ipynb');
I dont see any graphs in html report. It has been quite a task to figure out what's happening.
Same issue here: trying to use hbox and display to have a pandas df and a matplotlib.pyplot figure side-by-side: works like a charm on the notebook: really beatiful. It's empty when exported ...
display(Markdown(f'### {title}'), clear=False)
# plot and table
widget1 = widgets.Output()
widget2 = widgets.Output()
# render in output widgets
with widget1:
display(pandas_table)
with widget2:
display(plt_fig)
# add some CSS styles to distribute free space
box_layout = Layout(
display='flex',
flex_flow='row',
justify_content='space-around',
width='auto'
)
# create Horisontal Box container
hbox = widgets.HBox([widget1, widget2], layout=box_layout)
# render hbox
display(hbox)
hello, any updates here? I've experienced the same issue as well ...
Hello, any update ? I am using the same code as erickfis but to display 2 pandas df. Displays perfectly on the notebook but when converting I get 2 columns displaying the HTML code of the tables. using nbconvert 7.2.6
I am also having this problem. NBConvert 6.5.3
I need help with NBconvert 7.2.6. For example, I try to export HTML display widgets with the code below but retrieve
HTML(value='\n<svg width="280" height="280">\n <path d="M 265.0 155.0 L 253.545602565321 201.47231720437685…. Also, interactive widgets are not working.
import numpy as np
import ipywidgets as widgets
from IPython.display import display
import math
# create a range of values from 0 to 14
m=13
values = np.arange(m-1)
# define the radius of the circle
radius = 100
radius2=110
t=""
# calculate the coordinates of the points on the circle
points = []
for i in range(m):
angle = 2 * math.pi * i / m
x = 55+radius2 + radius * math.cos(angle)
y = 45+radius2 + radius * math.sin(angle)
points.append((x, y))
t+='<text x="'+str(50+radius2 + radius2 * math.cos(angle))+'" y="'+str(50+radius2 + radius2 * math.sin(angle))+'" fill="black">'+str(i)+'</text>'
t+='<text x="'+str(50+radius2)+'" y="'+str(50+radius2)+'" fill="black">mod '+str(m)+'</text>'
# create the circle as a path element
path = "M " + str(points[0][0]) + " " + str(points[0][1])
for point in points[1:]:
path += " L " + str(point[0]) + " " + str(point[1])
path += " Z"
circle = widgets.widgets.HTML(f"""
<svg width="{2*radius+80}" height="{2*radius+80}">
<path d="{path}" fill="none" stroke="black" stroke-width="2"/>
{t}
</svg>
""")
# display the circle
display(circle)
Related posts?:
[] This is a known issue, but there does not seem to be any solution at this point other than using NBClassic. <> https://discourse.jupyter.org/t/reloading-widget-state-in-saved-jupyter-notebooks/21295
[]
The widget state is still there and saved by JLab. Opening the notebook in NBClassic running against the Jupyter Server 2.7.0 shows the widget states and correctly indicates broken connections for widgets that talk to the kernel. Somehow JLab/Notebook 7 does not find the widget states in Notebooks even though it saves them. <> https://github.com/jupyter-widgets/ipywidgets/issues/3758
[] From my testing ipywidgets>=7.7, <8 works with Lab 3.6.6 and has correct state restore.
Jupyter Lab 4 requires ipywidgets>=8, in which I have found state restore is broken. <> https://github.com/jupyterlab/jupyterlab/issues/15361
update, related post (for vscode):
HTML ipywidgets state is not saved with the notebook - Stack Overflow https://stackoverflow.com/questions/37542298/html-ipywidgets-state-is-not-saved-with-the-notebook
Save notebook widget state in vs code - Stack Overflow https://stackoverflow.com/questions/65735851/save-notebook-widget-state-in-vs-code
Save notebook widget state · Issue #4404 · microsoft/vscode-jupyter https://github.com/microsoft/vscode-jupyter/issues/4404