xMIP
 xMIP copied to clipboard
xMIP copied to clipboard
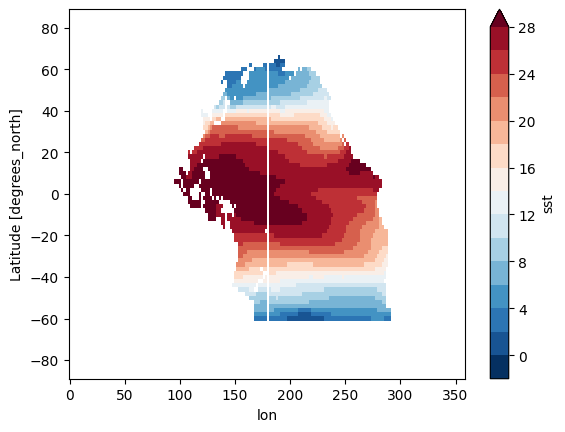
The stripe emerged after masking the ERSSTv5 SST data
after i tried to mask the Pacifca ocean in ERSSTv5 data, the stripe emerged
import xarray as xr
from xmip.regionmask import merged_mask
import regionmask
import numpy as np
basins = regionmask.defined_regions.natural_earth_v4_1_0.ocean_basins_50
sst = xr.open_dataset(cfg.Data_Path["ERSSTV5"]).sst
mask = merged_mask(basins,sst)
sst_masked = sst.where(np.logical_or(np.logical_or(mask == 2, mask==3),mask==4))
sst_masked.mean(dim = 'time').plot(levels = np.arange(-2,30,2))
Hi @jwy-wbe thanks for using xMIP, and sorry for the delayed response. That is certainly not ideal. Is the dataset you are using available publicly? I am currently not able to reproduce this line
sst = xr.open_dataset(cfg.Data_Path["ERSSTV5"]).sst
Hi @jwy-wbe thanks for using xMIP, and sorry for the delayed response. That is certainly not ideal. Is the dataset you are using available publicly? I am currently not able to reproduce this line
sst = xr.open_dataset(cfg.Data_Path["ERSSTV5"]).sst
I appreciate you taking the time to reply. The dataset is NOAA Extended Reconstructed SST V5 (A global monthly SST analysis from 1854 to the presen,2.0 degree latitude x 2.0 degree longitude global grid (89x180)) frequently used in scientific research, and it can be downloaded through this website (https://psl.noaa.gov/data/gridded/data.noaa.ersst.v5.html).
I think the problem might be the longitude of dataset is 0.0E - 358.0E which contains the 180.0E, and the maks values fall in 180E are all NaN. So how can I fix the maks values in 180E to make sure the plot are complete like this
image.

The figure comes from the paper "Disentangling GlobalWarming, Multidecadal Variability,and El Niño in Pacific Temperatures" published on Geophysical Research Letters
I think this might be an issue related to regionmask, but I am not 100% sure. To narrow this down I suggest the following:
- Define a much simpler mask that covers most of the North Pacific, like a lat/lon box. Does the problem still occur in this case? Then I would assume that this is a problem related to the regionmask package and you could raise an issue over there.
- If that does not show the problem there might be an issue with the masks defined in the natural_earth_v4_1_0 data.
I hope that can shed some light on the problem
I think this might be an issue related to regionmask, but I am not 100% sure. To narrow this down I suggest the following:
- Define a much simpler mask that covers most of the North Pacific, like a lat/lon box. Does the problem still occur in this case? Then I would assume that this is a problem related to the regionmask package and you could raise an issue over there.
- If that does not show the problem there might be an issue with the masks defined in the natural_earth_v4_1_0 data.
I hope that can shed some light on the problem
Thanks for your suggestion, I will try it later.
I enter the same problem. When I use a smaller subste data covering the North Pacific (100E-100W, 30-60N), the white line still exists.

Meanwhile, when I use "natural_earth_v5_0_0" data, the "merged_mask" function seems not work.
here is my code `import numpy as np import xarray as xr from xmip.regionmask import merged_mask import regionmask
fname = '/home/liuc/work/data/ocean/sst/ersstv5.nc' ds = xr.open_dataset(fname) basins = regionmask.defined_regions.natural_earth_v5_0_0.ocean_basins_50 mask = merged_mask(basins, ds) `
here is the error: File "/home/liuc/miniconda3/lib/python3.9/site-packages/xmip/regionmask.py", line 207, in merged_mask sb_idx = basins.map_keys(sb) File "/home/liuc/miniconda3/lib/python3.9/site-packages/regionmask/core/regions.py", line 166, in map_keys key = self.region_ids[key] KeyError: 'Ross Sea Eastern Basin'
version of regionmask and xmip regionmask == '0.9.0' xmip == '0.6.1rc0'
I enter the same problem. When I use a smaller subste data covering the North Pacific (100E-100W, 30-60N), the white line still exists.
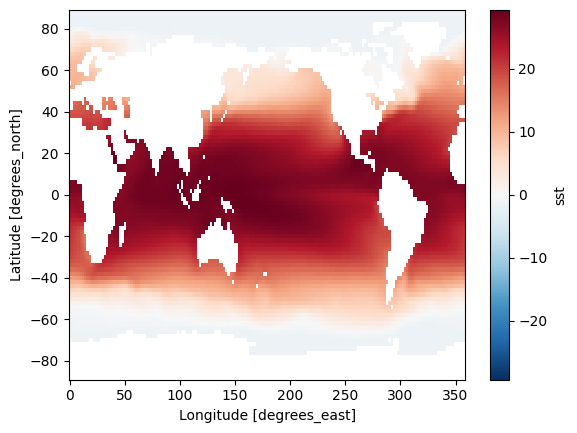
I fix the problem by using the mask values next to the 180 line (for example 177.5E), it turns out like this
I enter the same problem. When I use a smaller subste data covering the North Pacific (100E-100W, 30-60N), the white line still exists.
I fix the problem by using the mask values next to the 180 line (for example 177.5E), it turns out like this
Thank you for your remedy advice, now I can get it work!
Also, I still hope code contributors can help fix this bug anyway, if there is indeed a solution. @jbusecke
Meanwhile, when I use "natural_earth_v5_0_0" data, the "merged_mask" function seems not work.
here is my code `import numpy as np import xarray as xr from xmip.regionmask import merged_mask import regionmask
fname = '/home/liuc/work/data/ocean/sst/ersstv5.nc' ds = xr.open_dataset(fname) basins = regionmask.defined_regions.natural_earth_v5_0_0.ocean_basins_50 mask = merged_mask(basins, ds) `
here is the error: File "/home/liuc/miniconda3/lib/python3.9/site-packages/xmip/regionmask.py", line 207, in merged_mask sb_idx = basins.map_keys(sb) File "/home/liuc/miniconda3/lib/python3.9/site-packages/regionmask/core/regions.py", line 166, in map_keys key = self.region_ids[key] KeyError: 'Ross Sea Eastern Basin'
version of regionmask and xmip regionmask == '0.9.0' xmip == '0.6.1rc0'
Yeah this is a problem that we have discussed upstream. The addition of ocean basins to natural earth would make the functionality here obsolete (you could just use regionmask+natural earth, without the need for xmip).
I fix the problem by using the mask values next to the 180 line (for example 177.5E), it turns out like this
I do not quite understand how this fix works, and would be curious to know. Can you past the code that fixed the issue?
Meanwhile, when I use "natural_earth_v5_0_0" data, the "merged_mask" function seems not work. here is my code
import numpy as np import xarray as xr from xmip.regionmask import merged_mask import regionmask fname = '/home/liuc/work/data/ocean/sst/ersstv5.nc' ds = xr.open_dataset(fname) basins = regionmask.defined_regions.natural_earth_v5_0_0.ocean_basins_50 mask = merged_mask(basins, ds)here is the error: File "/home/liuc/miniconda3/lib/python3.9/site-packages/xmip/regionmask.py", line 207, in merged_mask sb_idx = basins.map_keys(sb) File "/home/liuc/miniconda3/lib/python3.9/site-packages/regionmask/core/regions.py", line 166, in map_keys key = self.region_ids[key] KeyError: 'Ross Sea Eastern Basin' version of regionmask and xmip regionmask == '0.9.0' xmip == '0.6.1rc0'Yeah this is a problem that we have discussed upstream. The addition of ocean basins to natural earth would make the functionality here obsolete (you could just use regionmask+natural earth, without the need for xmip).
I fix the problem by using the mask values next to the 180 line (for example 177.5E), it turns out like this
I do not quite understand how this fix works, and would be curious to know. Can you past the code that fixed the issue?
Here is my code to fix the issue. It works on ERSSTV5 SST dataset which I mentioned before. I am not sure this method will work on other dataset. The key line is basin_mask.loc[dict(lon = 180)]=basin_mask.sel(lon = 180 - np.abs(mask.lon.diff(dim = 'lon')[0])). The idea is to use the mask value next to the 180 degree fill the Nan of 180 degree. But I am not sure this is correct.
def mask_Ocean_basin(ds,basin = 'pacifica',standard = 'xmip'):
if standard == 'ar6':
region = {
"Pacific":("N.Pacific-Ocean","Equatorial.Pacific-Ocean","S.Pacific-Ocean"),
"Atlantic":("N.Atlantic-Ocean","Equatorial.Atlantic-Ocean","S.Atlantic-Ocean"),
"Indian":("Equatorial.Indic-Ocean","S.Indic-Ocean"),
}
ocean_basins = regionmask.defined_regions.ar6.all
mask = ocean_basins.mask(ds.lon,ds.lat)
mask_values = [ocean_basins.region_ids[regin] for regin in region[basin.title()]]
elif standard == 'xmip':
ocean_basins = regionmask.defined_regions.natural_earth_v4_1_0.ocean_basins_50
mask = merged_mask(ocean_basins,ds)
region = {
"Pacific":(2,3,4),
"Atlantic":(0,1),
"Indian":(5),
}
mask_values = [value for value in region[basin.title()]]
if len(mask_values) == 3:
basin_mask = (mask == mask_values[0]) | (mask == mask_values[1]) | (mask == mask_values[2])
elif len(mask_values) == 2:
basin_mask = (mask == mask_values[0]) | (mask == mask_values[1])
else:
basin_mask = (mask == mask_values[0])
if basin.lower() == 'pacific':
### in case of the data does not have the 180 degree line
try:
basin_mask.loc[dict(lon = 180)]=basin_mask.sel(lon = 180 - np.abs(mask.lon.diff(dim = 'lon')[0]))
except:
basin_mask = basin_mask
### update the Attributes
attrs = ds.attrs
attrs['basin_masked'] = f"Maks the other basin except {basin.title()}, based on the {standard.upper()} standard"
ds_masked = ds.where(basin_mask)
return ds_masked.assign_attrs(attrs)
Ah thank you. So you are saying that one column of the lat values are nan? that would absolutely explain this issue. Do you think we can close this issue now?
Ah thank you. So you are saying that one column of the lat values are nan? that would absolutely explain this issue. Do you think we can close this issue now?
Yes,thanks you.
