pyxem
 pyxem copied to clipboard
pyxem copied to clipboard
Proper Dealing with Masks
This was something that I was meaning to bring up the other day during the developers meeting but forgot. I think that is deserves some discussion possibly here and potentially upstream in hyperspy. Let me know if I should move the discussion there, I opened an issue request there a little while ago but never got around to submitting a pull request due to problems with masking and lazy signals.
I will try and describe the different masking options available and maybe we can think about standardizing them.
Hyperspy
- hyperspy supports using numpy.ma.masked_arrays as the data variable for some signal class. - For lazy signals the dask.ma class is very easily interchangeable but there is an issue that dask arrays are immutable and so either the dask array must be copied and changed or the mask must be set using a non lazy signal and then cast to a lazy one which kind of defeats the purpose
- I am not sure if hyperspy deals with saving masked data arrays properly
- Hyperspy also has a fairly developed roi module which works similar to a mask but doesn't have the flexibility of one.
Empyer
- In dealing with masks I have played with two different options:
- The first is forgoing masking on lazy signals and using hyper spy.
- The second is creating two different masks for the signal and the navigation axes. This saves on memory a fair amount but doesn't give you quite as much freedom with creating masks.
- In both cases, a useful feature I have implemented is the ability to slice and then apply a mask. For example if I wanted to mask a circle on only one pattern at positions [3,4] I could do
signal.manav[3,4].mask_circle(center=(0.,0.), radius=1.0)Basically this just copies a lot of the functionality of the Special slicer class in hyper spy.
PixSTEM @magnunor I don't exactly know how you deal with masks but I think that you have a couple of methods dealing with editing masks using Dask? It also seems like you pass in masks as boolean arrays for your functions?
PyXEM
- For the most part pyXEM defines masks as either 2-d boolean arrays (of the same size as the signal) to be passed in as an argument for a methods.
- Or as 2-D boolean Signals of the same shape as the as the signal to be iterated over.
Ideal Solution? I think the eventually the goal would be to add masking support upstream to hyperspy. Using the NumPy and dask masking capabilities would probably work due to the flexibility and existing functions like nan-mean, nan-sum etc.. It would add on to the memory required for some data set but nothing overly large.
The question that I have is what is the best way to change the mask array with a lazy signal?
Beyond that I am partial to the idea of adding a new set of SpecialSlicer objects masig and manav which work as follows:
signal.masig[0:10,:]=True # masks pixels from pixel 0 to pixel 10 over all navigation axes
signal.masig[0.:10.,:]=True # masks from 0.0 to 10.0 using real units over all navigation axes
signal.masig[0.:10.,:]=False #unmasks from 0.0 to 10.0 using real units over all navigation axes
signal.manav[1,1].masig[0.:10.,:]=True # masks from 0.0 to 10.0 using real units on pattern (1,1)
signal.manav[1,1].mask_circle(center=(10.,10.), rad=1.) # masks a circle on pattern (1,1) with center (10.0,10.0) and radius 1.0 in real units.
signal.mask_circle(center=(10.,10.), rad=1.) # masks a circle on all patterns with center (10.0,10.0) and radius 1.0 in real units.
signal.mask_circle(center=(10,10), rad=10) # masks a circle on all patterns with center (10,10) and radius 10 in pixels
After looking into this further I realized that the best way to handle this might be to work more in the lines of what is already implemented in hyperspy.
The slicing in Hyperspy is already well implemented so all you need is a method to change the data type from numpy array to a masked array. Once the data is a masked array, you can just set the array as s.isig[:,1:5]= np.ma.masked
Additionally because of how Hyperspy's slicing works you can also use a ROI for slicing.
So it looks like this is something that isn't going to get into Hyperspy any time soon. The issue is that the numpy.ma (and and the Dask.ma class) which seem to be at odds with each other.
Right now in the diffraction2d class we have a couple of methods which deal with masking:
https://github.com/pyxem/pyxem/blob/9635102bb34e1b08e343379ed3a11ce21ab08731/pyxem/signals/diffraction2d.py#L325
https://github.com/pyxem/pyxem/blob/9635102bb34e1b08e343379ed3a11ce21ab08731/pyxem/signals/diffraction2d.py#L444
https://github.com/pyxem/pyxem/blob/9635102bb34e1b08e343379ed3a11ce21ab08731/pyxem/signals/diffraction2d.py#L504
https://github.com/pyxem/pyxem/blob/9635102bb34e1b08e343379ed3a11ce21ab08731/pyxem/signals/diffraction2d.py#L849
https://github.com/pyxem/pyxem/blob/9635102bb34e1b08e343379ed3a11ce21ab08731/pyxem/signals/diffraction2d.py#L1611
And I think that they have a variety of places we could improve:
- None of them allow for using "real units" like hyper spy does. This makes it more difficult to create a mask quickly based on what you already know about the image or it makes it harder to say mask anything below 1 nm^-1 etc.
- There is no mask saved beside the diffraction pattern. This is something that I need to add into hyperspy. But maybe it deserves some discussion. Should have 1 mask applied to every x, y or a mask which is the same size as the data. The first one is easier, speeds up computations sometimes but it isn't the greatest for flexibility
- Hyperspy has some support for masking we could build on but that might be a little faulty. I'll see what I can get in to Hyperspy and then move from there
I've been thinking about what we're likely to want in 0.14 recently, and I think this high (near top) on that agenda. Can I get back to you on specifics?
Yea, I can clean up the code that I was going to try to get into hyperspy and see if they still want to merge it. I think that for the most part that allows for using ROI's to create masks and allows masks to be saved. I think that is a good starting point if we want to create a more intuitive and flexible set of masks in pyxem.
I'd like to stay in the loop on this as I've been dealing with masking myself but not in the most sustainable way. My use case was creating a mask out of diffsims patterns from an orientation map (each diffraction spot gets mapped to a true or false circle), so I have a different mask for each pattern in the map. @CSSFrancis I guess something like this is in line with one of the cases you describe above? I use these masks then to create a VDF that excludes all the indexed spots to highlight unindexed phases. I might add my mask creation process into diffsims. Now my VDF creation procedure just happens via multiplication using map_blocks and masks generated on the fly for each pattern. A persistent lazy masking operation is much more attractive.
Lately, I've been thinking whether it would not make sense to implement masks as a separate class, perhaps as a subclass of Signal2D. The advantage of this scenario is that building up a custom mask could be implemented via methods of the mask class, e.g. mask.add_circle, mask.add_poligon, mask.add_line, mask.add_array. etc. and it would be easy to add positive and negative elements. For example, a donut pattern to select a ring starts from a "false" array, then one adds a "true" circle and then again a smaller "false" circle. For common cases like the donut one can have shortcut methods of course, but I once needed a thin rotated rectangle to select the region between two diffraction spots:
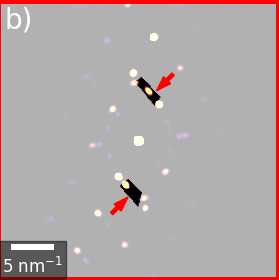 Then Signal2D could just have an
Then Signal2D could just have an apply_mask method, which internally masks the dask or numpy array.
But I wouldn't know how to do this if the mask is a 4D dask dataset itself.
Secondly, how would one generate a 4D mask if dask arrays are immutable? Might get very hacky and totally not performant.
@din14970 I need to get a little bit further into the Dask masking functions but I don't really see why masked arrays can't be lazy. As long as they are smart about waiting until a chunk is calculated to calculate the mask then it should work side by side the dask array with no problems. Dask is pretty good at "faking" that it has done some calculations, and masks are pretty easy to fake because you know the size and datatype.
It wouldn't really be that hard to add in masking capabilities. The problem is that hyperspy wants masking to work like:
np.ma.mask_below(signal, 0)
Where the signal acts like a numpy array. This actually works for just about any numpy signal, expect for masked arrays which are super problematic because they are an older legacy class in numpy and as a result they have special permissions which wreaks everything. It is kind of a rabbit hole I would not recommend going down.
The work around is fairly simple, just making a pyxem.ma module that just wraps the da.ma and np.ma classes and maybe adds in some other methods. I can add that in pretty easily when I get the chance
A tangentially related note on masks which I thought I would leave here, I've added some mask creation utilities to diffsims here.
In addition, I've found a rough but O.K. solution to my own problem of creating a lazy 4D mask the same size as the dataset from an orientation indexing result. In the method below, dataset is a dask array representing the 4D STEM data, results represents the output (a dictionary) from index_dataset_with_template_rotation, and library_sims represents the list of calculated templates. I was struggling quite a bit with how one should generate a dask array from scratch, since assignment in dask arrays is not supported; with map blocks and using a "block-aware" function this can be achieved.
def get_mask_dataset_2(dataset, results, library_sims, phase=None, radius=8, *args, **kwargs):
"""Return a dask array of the same shape and chunks as the dataset with corresponding masks"""
if phase is None:
phase = list(results.keys())[0] # select the first phase if nothing is provided
indexes = results[phase]["template_index"][:,:,0] # index in list of simulations corresponding to best template
orientations = results[phase]["orientation"][:,:,0,:] # orientation of best fit template
# create a dask array of masks
imshap = dataset.shape[-2:]
def create_mask_block(block_info=None):
loc = block_info[None]["array-location"] # boundaries of the block in the array
dtyp = block_info[None]["dtype"]
bby = loc[0][1] - loc[0][0] # size of the block
bbx = loc[1][1] - loc[1][0]
sims = np.empty((bby, bbx, imshap[0], imshap[1]), dtype=dtyp)
for r in range(bby):
abs_y = loc[0][0] + r
for c in range(bbx):
abs_x = loc[1][0] + c
in_plane_angle = orientations[abs_y, abs_x, 0]
simulation = library_sims[indexes[abs_y, abs_x]]
sims[r, c] = simulation.get_as_mask(imshap, radius, True, None, None, in_plane_angle)
return sims
masks = da.map_blocks(
create_mask_block,
chunks = dataset.chunks,
dtype=bool,
)
return masks
Initially I was working with dask.delayed for creating the masks from the templates but then I had to store these in a list, which I needed to stack and reshape. This meant that computing a single pattern took forever because dask didn't know how to parallelize it properly. The above implementation works relatively efficiently and can give me a mask from a specific location rather quickly. The 4D mask can then be applied to the 4D dataset simply with
masked_data = da.ma.masked_where(lazy_masks, lazy_data)
Semi-related: there is some support for this in dask_tools.py: https://github.com/pyxem/pyxem/blob/master/pyxem/utils/dask_tools.py#L403, which is for example used for doing center_of_mass.
use #727 for this.