openff-toolkit
 openff-toolkit copied to clipboard
openff-toolkit copied to clipboard
[REQUEST] Diagnostic regression tests
Is your feature request related to a problem? Please describe.
Some time ago, the charge model changed from ELF10 to single-conformer AM1BCC without many people noticing, possibly unintentionally. This seems dangerous.
Describe the solution you'd like
A regression test that tests for specific "default" behaviour. I know this does not currently exist because I changed the charge model back to ELF10 and all tests passed. That means, if the model should change again, even those checking over the PR may not be fully aware. https://github.com/openforcefield/openff-toolkit/commit/60ce9594a7f79b766d1359809c9f52163abdbcae
Describe alternatives you've considered
Perhaps the absence of failing tests means that the actual charge differences are negligible, at least in the test cases currently in the test suite. However, it would still be good for any changes to the default settings to require some change to the tests.
Additional context
actual charge differences [might be] negligible
From your tinkering you have a sense of what order of magnitude these differences tend to be?
FTR we added some basic energy tests in https://github.com/openforcefield/openff-toolkit/commit/14c963fae0bc2e12d328070eb726a7848f87cf25 as part of the saga resolved in #808. This happened before #722; it's possible but not guaranteed that these would have caught that change. I don't know if differences in partial charges would be enough to be picked up there.
The scope of test_energies.py is small, but should in principle catch a lot of behavior changes. Still, I'd strongly be in favor of expanding it to cover things it currently misses.
It looks like that should have been caught between 0.8.3. and 0.8.4? Or, put more precisely, it looks like there were differences in the energies generated by each version of the toolkit. I generated the JSON files by running the below script after switching versions via git tax vX.Y.Z.
{'0.7.0': {'CID20742535_anion.sdf_constrained': 276.1610283626506,
'CID20742535_anion.sdf_unconstrained': 276.2305291512185,
'ethanol.sdf_constrained': -10.218362843282843,
'ethanol.sdf_unconstrained': -10.198457704532483,
'methane_multiconformer.sdf_constrained': 16839.315152273277,
'methane_multiconformer.sdf_unconstrained': 16839.320757291953},
'0.8.3': {'CID20742535_anion.sdf_constrained': 276.1610283626506,
'CID20742535_anion.sdf_unconstrained': 276.2305291512185,
'ethanol.sdf_constrained': -10.218362843282843,
'ethanol.sdf_unconstrained': -10.198457704532483,
'methane_multiconformer.sdf_constrained': 2.0119485820433414,
'methane_multiconformer.sdf_unconstrained': 2.0175537970124706},
'0.8.4': {'CID20742535_anion.sdf_constrained': 276.1593481395032,
'CID20742535_anion.sdf_unconstrained': 276.2288489280711,
'ethanol.sdf_constrained': -10.218464321789973,
'ethanol.sdf_unconstrained': -10.198559183039613,
'methane_multiconformer.sdf_constrained': 2.0119485820433414,
'methane_multiconformer.sdf_unconstrained': 2.0175537970124706}}
0.00010147850712982631
0.00010147850712982631
0.0
0.0
0.001680223147388915
0.001680223147388915
https://gist.github.com/mattwthompson/fedfef08cf5728080a10f96023919c23
From your tinkering you have a sense of what order of magnitude these differences tend to be?
Erm, kind of. On a very specific dataset (Ace-Val-X-Y-Z-Val-Nme capped pentapeptides), mostly < 0.1 e. But that comes with caveats like only examining 1 conformer for AM1BCC instead of trying a few different ones, and I think that's a little larger than the typical molecule in the Parsley dataset. (Y is not tyrosine, just another placeholder for any amino acid). I'm still trying to figure out how high a magnitude of difference affects the energies significantly, though.
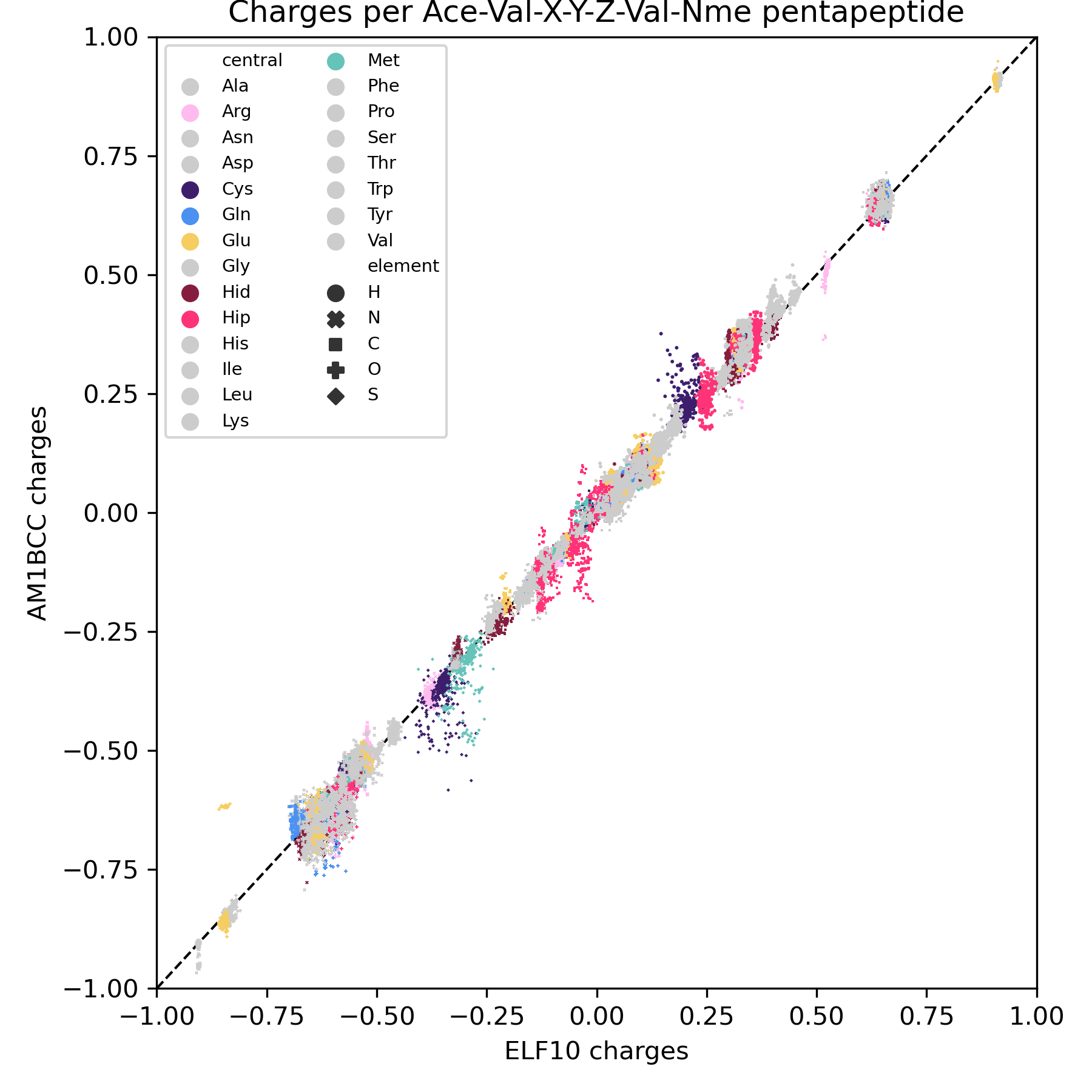
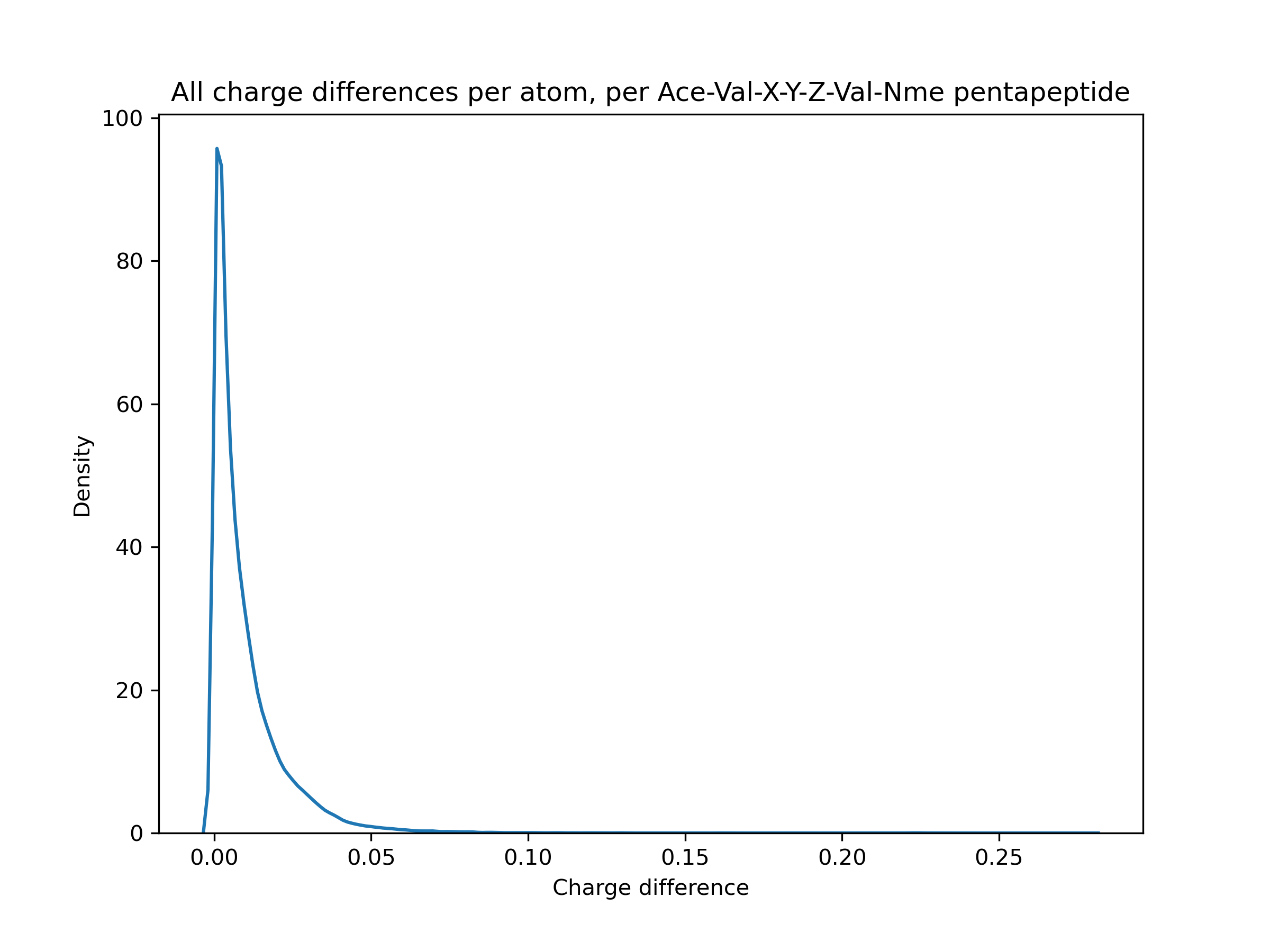
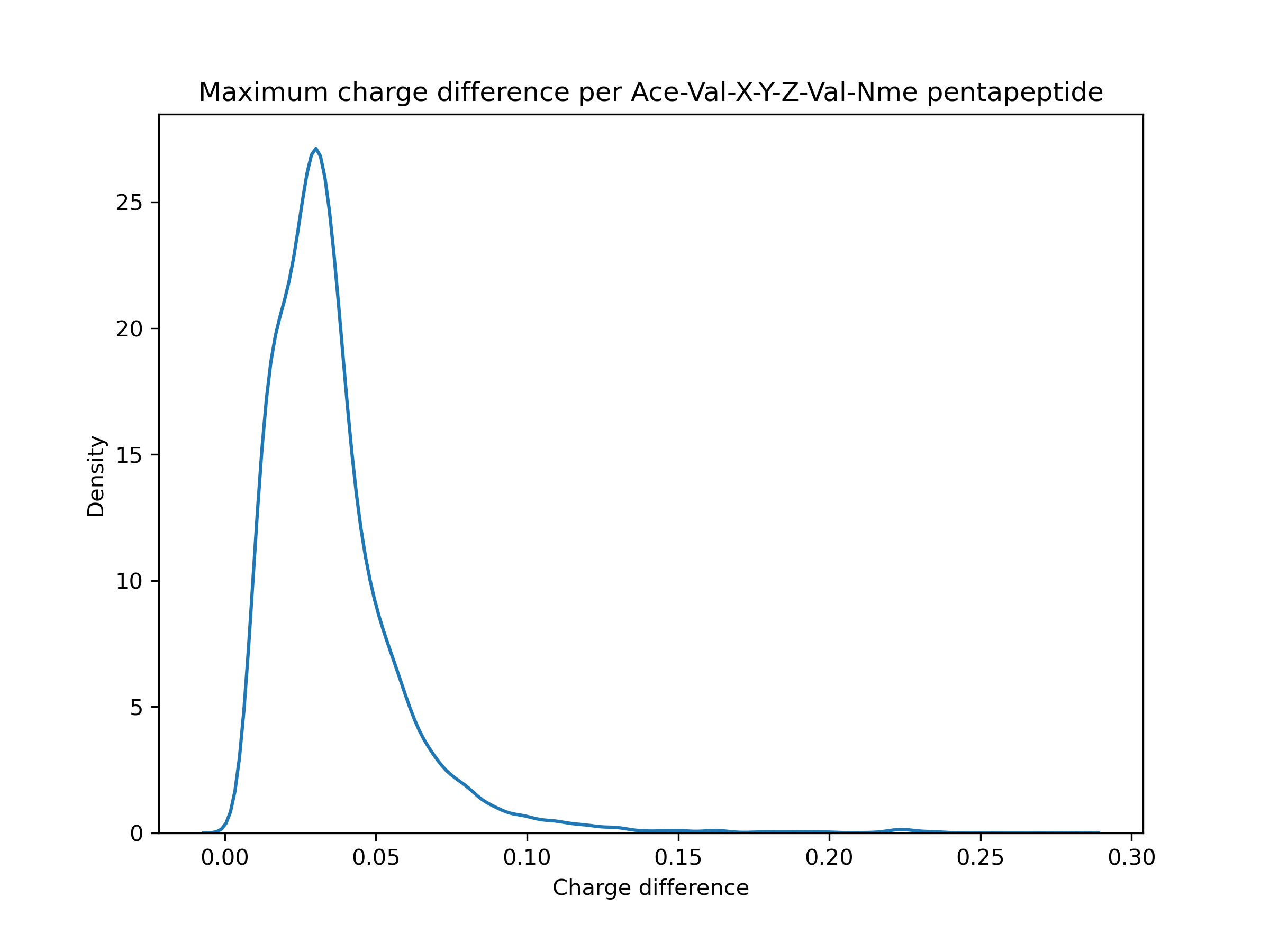
It looks like that should have been caught between 0.8.3 and 0.8.4?
I am a little confused, #722 is tagged to be in the 0.9.1 release. Was it actually introduced in 0.8.3 or 0.8.4 ?
I don't have a great feel for the lower bound of how much run-to-run/version-to-version variance is generally acceptable (mean errors of 0.1 are probably too much but tightening things to smaller errors like 0.000001 would be silly, given the other errors in MM workflows) - especially given headaches like #1019 - but my general impression here is that these errors are quite a bit bigger than we should allow silently happen. Sure, most are like 0.001 - 0.02, but there's a tail > 0.05 and even a little bump around 0.22. That seems significant.
_ Was it actually introduced in 0.8.3 or 0.8.4 ?
Those milestones tend to be forward-looking, not merged into the releases they're planned on, and forgotten about after merging. I tried to infer from the dates when it was merged and when releases were made ... but I clearly did something wrong. Probably forgetting that 0.8.4 was not mainline.
This S/O copy-paste says the same thing:
$ git tag --contains 2380061f117ef75e7b565a4fe413e746650f6cdf | cat
0.10.0
0.9.1
0.9.2
latest
So here's the same bit comparing 0.8.3 and 0.9.1:
{'0.8.3': {'CID20742535_anion.sdf_constrained': 276.1610283626506,
'CID20742535_anion.sdf_unconstrained': 276.2305291512185,
'ethanol.sdf_constrained': -10.218362843282843,
'ethanol.sdf_unconstrained': -10.198457704532483,
'methane_multiconformer.sdf_constrained': 2.0119485820433414,
'methane_multiconformer.sdf_unconstrained': 2.0175537970124706},
'0.9.1': {'CID20742535_anion.sdf_constrained': 276.1593481395032,
'CID20742535_anion.sdf_unconstrained': 276.2288489280711,
'ethanol.sdf_constrained': -10.218464321789973,
'ethanol.sdf_unconstrained': -10.198559183039613,
'methane_multiconformer.sdf_constrained': 2.0119485820433414,
'methane_multiconformer.sdf_unconstrained': 2.0175537970124706}}
0.00010147850712982631
0.00010147850712982631
0.0
0.0
0.001680223147388915
0.001680223147388915
These (0.9.1 vs 0.8.3) are the same numbers as 0.8.4 vs. 0.8.3, which reminds me that 0.8.4 and 0.9.1 were meant to be functionally equivalent but with the old imports in order to not need to refactor the benchmarking code. Maybe I need more coffee before doing diagnostic work ...
I strongly agree that we want diagnostic regression tests, such as those that would have caught this. Certainly sudden changes in charges of 0.01 or 0.1 e ought to result in issues, or energy changes of a couple of tenths of a kcal/mol ought to be caught.
Some code changes will perhaps result in larger deviations than this that are OK, but we would want to know about them and assess, meaning that we really ought to have somehting in place which is flagging them for us.
I'm working on a set of regression tests in another repo, which is a part of the Interchange adoption plan. Despite the naming, it's toolkit-centric and meant to test results from different conda environments, presumably with different versions of the toolkit installed. It's a work in progress and will change a few times before we want to have much faith in it, but once online it could be molded into a pre-release turnkey. A notable difference is that is mostly relies on comparing serialized representations of the OpenMM Systems; this should generally be a higher standard to meet than energy differences but there may be subtle differences between the approaches.
A minor annoyance is that because it's meant to test different versions of the toolkit, it compares against different Python calls, it absorbs the problems of run-to-run variance of wrapped methods. This is mostly AM1-BCC charges but can sometimes affect other things like fractional bond orders.
It might be useful to put some bounds on what behavior we really must avoid regressions with, or at least split things up. If it's a matter of the charges/conformers/etc. that result from various Molecule API calls, that'll be hard for us to take responsibility for given the number of ways things may differ run-to-run or with small changes upstream. Results from OpenMM system creation can get dubious depending on the standards that are used ... we put a good bit of work into https://github.com/openforcefield/interchange-regression-testing and I'm currently trying to add automation to that repo and a subset of those tests in the Interchange test suite. "Default" behavior is hard to get a handle on given the number of things that users might do, and there's also the question of "default behavior of what?"
From the other perspective, we could build things from the ground up one by one. That list of tests would be curated first from issues we've had in the past and then expanded to things we're generally worried about.