nipype
 nipype copied to clipboard
nipype copied to clipboard
Compcor
Hi, I am having issues running Compcor on some of my fmri sessions. I get the following error on 2 out of 10 days of scanning only, in all runs of those days. I have visually checked all my input files, like functional scans, and WM/CSF masks, but I can't seem to find anything blatantly wrong when compared with the successful sessions, at least from my inexpert eye (that's my first time with Compcor!). The txt file "acompcor" is created but it's completely empty, (I have a number of rows equal to the volumes in my scan, but in this case they're filled with nothing). Therefore the code stops when it gets to calculate tcompcor from acompcor, saying that it
cannot do a non-empty take from an empty axes.
Does anyone have had similar issues? Any solution &/or feedback would be greatly appreciated!!
This is the error I get after running it:
Traceback (most recent call last):
File "/usr/lib/python3/dist-packages/thonny/backend.py", line 1213, in wrapper
result = method(self, *args, **kwargs)
File "/usr/lib/python3/dist-packages/thonny/backend.py", line 1200, in wrapper
return method(self, *args, **kwargs)
File "/usr/lib/python3/dist-packages/thonny/backend.py", line 1272, in _execute_prepared_user_code
exec(statements, global_vars)
File "/mnt/Seagate_Expansion_Drive/fmri_monty/Raw/CompCors11.py", line 58, in <module>
func_CompCor(filepath,subdir,f'run{tail}')
File "/mnt/Seagate_Expansion_Drive/fmri_monty/Raw/CompCors11.py", line 48, in func_CompCor
tCompCor.run()
File "/home/irecap/.local/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 434, in run
runtime = self._run_interface(runtime)
File "/home/irecap/.local/lib/python3.8/site-packages/nipype/algorithms/confounds.py", line 628, in _run_interface
mask_images = self._process_masks(mask_images, imgseries.dataobj)
File "/home/irecap/.local/lib/python3.8/site-packages/nipype/algorithms/confounds.py", line 842, in _process_masks
threshold_std = np.percentile(
File "<__array_function__ internals>", line 5, in percentile
File "/home/irecap/.local/lib/python3.8/site-packages/numpy/lib/function_base.py", line 3732, in percentile
return _quantile_unchecked(
File "/home/irecap/.local/lib/python3.8/site-packages/numpy/lib/function_base.py", line 3851, in _quantile_unchecked
r, k = _ureduce(a, func=_quantile_ureduce_func, q=q, axis=axis, out=out,
File "/home/irecap/.local/lib/python3.8/site-packages/numpy/lib/function_base.py", line 3429, in _ureduce
r = func(a, **kwargs)
File "/home/irecap/.local/lib/python3.8/site-packages/numpy/lib/function_base.py", line 3967, in _quantile_ureduce_func
x1 = take(ap, indices_below, axis=axis) * weights_below
File "<__array_function__ internals>", line 5, in take
File "/home/irecap/.local/lib/python3.8/site-packages/numpy/core/fromnumeric.py", line 191, in take
return _wrapfunc(a, 'take', indices, axis=axis, out=out, mode=mode)
File "/home/irecap/.local/lib/python3.8/site-packages/numpy/core/fromnumeric.py", line 58, in _wrapfunc
return bound(*args, **kwds)
IndexError: cannot do a non-empty take from an empty axes.
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/usr/lib/python3/dist-packages/thonny/backend.py", line 1259, in execute_source
return self._execute_prepared_user_code(statements, expression, global_vars)
File "/usr/lib/python3/dist-packages/thonny/backend.py", line 1218, in wrapper
return {"user_exception": self._vm._prepare_user_exception()}
File "/usr/lib/python3/dist-packages/thonny/backend.py", line 1027, in _prepare_user_exception
"stack": self._export_stack(last_frame),
File "/usr/lib/python3/dist-packages/thonny/backend.py", line 908, in _export_stack
module_name = system_frame.f_globals["__name__"]
KeyError: '__name__'
This is my code, which works for majority of the days.
# Import global engines
import os # system
from os.path import abspath
import multiprocessing
import nibabel as nib
import numpy as np
# Import numpy engines
import nipype.pipeline.engine as pe # the workflow and node wrappers
from nipype import Function
import nipype.interfaces.io as nio # Input/Output
import nipype.interfaces.utility as util # utility
import nipype.interfaces.fsl as fsl # fsl
import nipype.interfaces.afni as afni # afni
import nipype.interfaces.ants as ants # ANTS
import nipype.algorithms.confounds as conf #CompCor
# Import nilearn
from nilearn.masking import compute_epi_mask
TR = 2.12
aCompCor_varThresh = 0.5
tCompCor_varThresh = 0.5
def func_CompCor(inEPI, dir, run):
if not os.path.exists(f'{dir}/Preprocessed/CompCor'):
os.makedirs(f'{dir}/Preprocessed/CompCor')
aCompCor = conf.ACompCor(
realigned_file = inEPI,
mask_files = [f'{dir}/Preprocessed/b_Segmentation/{run}_CSF_funcspace_mask.nii.gz',f'{dir}/Preprocessed/b_Segmentation/{run}_WM_funcspace_mask.nii.gz'],
merge_method = 'union',
variance_threshold = aCompCor_varThresh,
repetition_time = TR,
pre_filter = 'cosine',
failure_mode = 'NaN',
components_file = f'{dir}/Preprocessed/CompCor/aCompCor.txt')
aCompCor.run()
tCompCor = conf.TCompCor(
realigned_file = inEPI,
mask_files = [f'{dir}/Preprocessed/b_Segmentation/{run}_CSF_funcspace_mask.nii.gz',f'{dir}/Preprocessed/b_Segmentation/{run}_WM_funcspace_mask.nii.gz'],
merge_method = 'union',
variance_threshold = tCompCor_varThresh,
repetition_time = TR,
pre_filter = 'cosine',
percentile_threshold=0.05,
failure_mode = 'NaN',
components_file = f'{dir}/Preprocessed/CompCor/tCompCor.txt')
tCompCor.run()
return
path = '/mnt/Seagate_Expansion_Drive/fmri_monty/Raw/s11'
for subdir, _, files in os.walk(path):
for filename in files:
filepath = subdir + os.sep + filename
(head, tail) = os.path.split(subdir)
if filepath.endswith('c_func.nii'):
print(filepath)
func_CompCor(filepath,subdir,f'run{tail}')
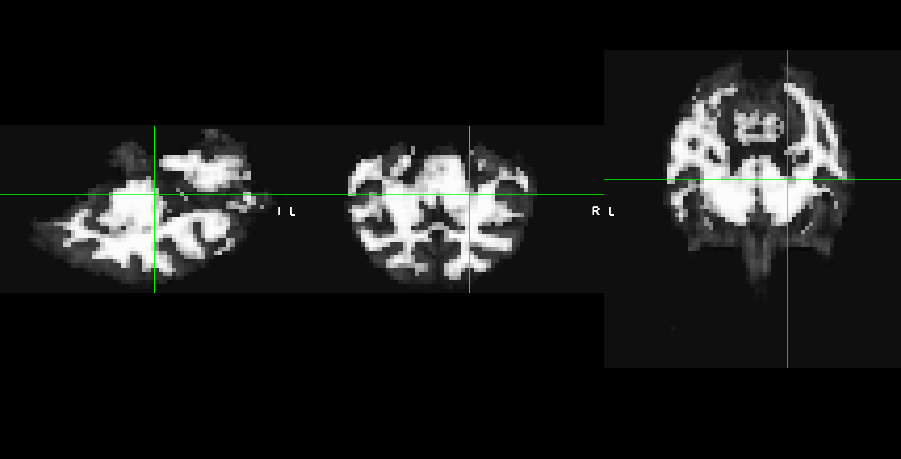
These are the CSF and WM masks respectively :
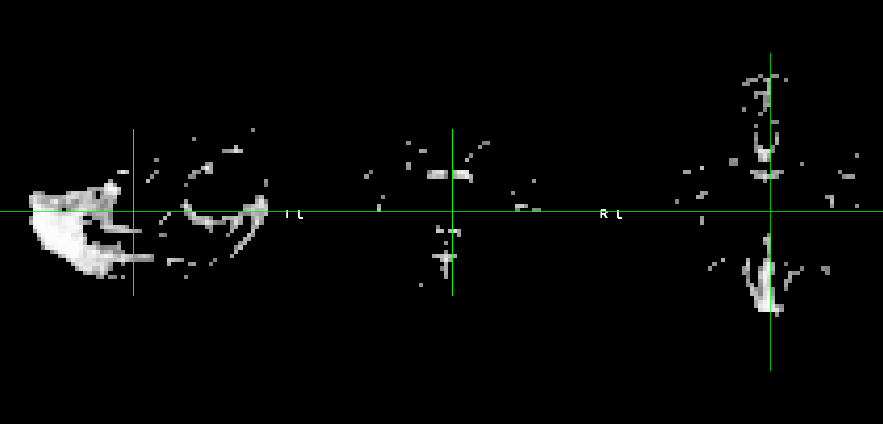
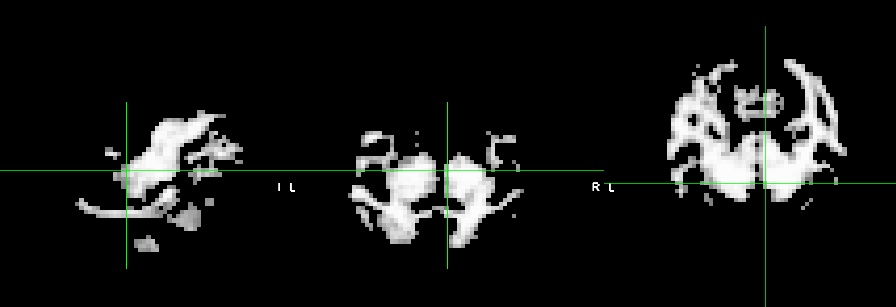
And this is the WM mask overlaied onto the functional of one run.