vscode-R
 vscode-R copied to clipboard
vscode-R copied to clipboard
Not all R Process (or radian process) will be killed when vscode exited in ubuntu
Describe the bug
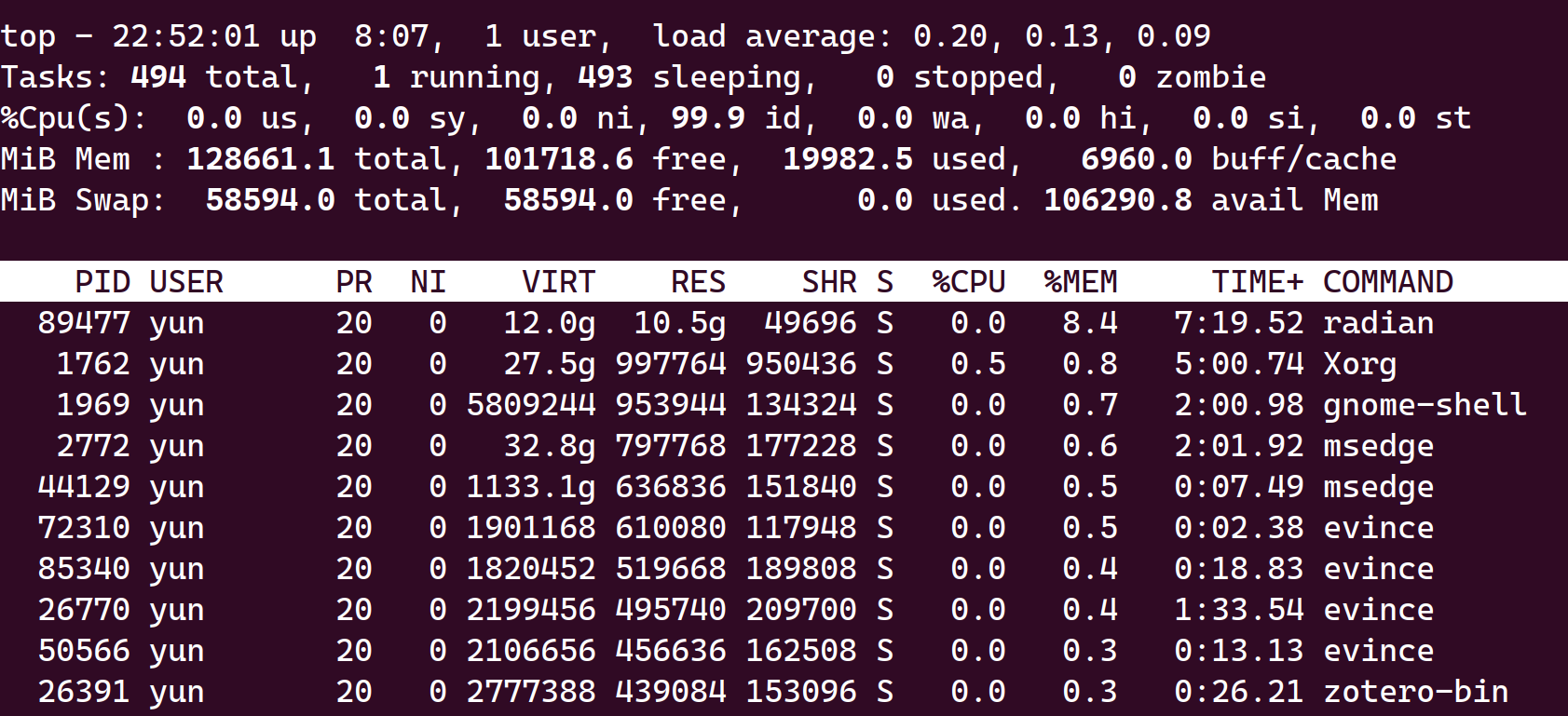
Not all R Process (or radian process) will be killed when vscode exited in ubuntu (22.04). See following picture where radian remains after vscode have been closed.
To Reproduce Sorry, I cannot reproduce this steadily. This issue has been reported in windows too. I have experienced twice in both situations I have used BiocParallel to implement parallelly computations.
Can you fix this issue by yourself? (We appreciate the help)
No, I have little experiences to write vscode extensions.
here is my sessioninfo to produce the issue:
[R]> sessionInfo()
R version 4.2.3 (2023-03-15)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.2 LTS
Matrix products: default
BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/libmkl_rt.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8
[4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=zh_CN.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=zh_CN.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] here_1.0.1 magrittr_2.0.3
[3] ggplot2_3.4.1 SingleCellExperiment_1.20.1
[5] SummarizedExperiment_1.28.0 Biobase_2.58.0
[7] GenomicRanges_1.50.2 GenomeInfoDb_1.34.9
[9] IRanges_2.32.0 S4Vectors_0.36.2
[11] BiocGenerics_0.44.0 MatrixGenerics_1.10.0
[13] matrixStats_0.63.0
loaded via a namespace (and not attached):
[1] colorspace_2.1-0 RcppEigen_0.3.3.9.3 rjson_0.2.21
[4] class_7.3-21 rprojroot_2.0.3 circlize_0.4.15
[7] scuttle_1.8.4 bluster_1.8.0 XVector_0.38.0
[10] RcppHNSW_0.4.1 GlobalOptions_0.1.2 BiocNeighbors_1.16.0
[13] clue_0.3-64 proxy_0.4-27 hexbin_1.28.3
[16] listenv_0.9.0 RSpectra_0.16-1 fansi_1.0.4
[19] ranger_0.14.1 codetools_0.2-19 sparseMatrixStats_1.11.1
[22] doParallel_1.0.17 cachem_1.0.7 robustbase_0.95-0
[25] jsonlite_1.8.4 ResidualMatrix_1.8.0 cluster_2.1.4
[28] png_0.1-8 compiler_4.2.3 ggplot.multistats_1.0.0
[31] dqrng_0.3.0 fastmap_1.1.1 SeuratObject_4.1.3
[34] Matrix_1.5-1 limma_3.54.2 cli_3.6.1
[37] BiocSingular_1.14.0 tools_4.2.3 rsvd_1.0.5
[40] igraph_1.4.1 gtable_0.3.3 glue_1.6.2
[43] GenomeInfoDbData_1.2.9 dplyr_1.1.1 ggthemes_4.2.4
[46] Rcpp_1.0.10 carData_3.0-5 vctrs_0.6.1
[49] progressr_0.13.0 iterators_1.0.14 DelayedMatrixStats_1.20.0
[52] scmisc_0.0.0.9000 lmtest_0.9-40 stringr_1.5.0
[55] laeken_0.5.2 globals_0.16.2 beachmat_2.14.0
[58] lifecycle_1.0.3 irlba_2.3.5.1 statmod_1.5.0
[61] future_1.32.0 edgeR_3.40.2 DEoptimR_1.0-11
[64] zoo_1.8-11 zlibbioc_1.44.0 MASS_7.3-58.3
[67] scales_1.2.1 VIM_6.2.2 pcaMethods_1.90.0
[70] parallel_4.2.3 RColorBrewer_1.1-3 qs_0.25.5
[73] curl_5.0.0 ComplexHeatmap_2.14.0 memoise_2.0.1
[76] stringi_1.7.12 foreach_1.5.2 ScaledMatrix_1.6.0
[79] e1071_1.7-13 destiny_3.12.0 TTR_0.24.3
[82] scran_1.27.3 boot_1.3-28 BiocParallel_1.32.6
[85] shape_1.4.6 rlang_1.1.0 pkgconfig_2.0.3
[88] bitops_1.0-7 lattice_0.20-45 purrr_1.0.1
[91] tidyselect_1.2.0 parallelly_1.35.0 R6_2.5.1
[94] generics_0.1.3 metapod_1.6.0 DelayedArray_0.24.0
[97] pillar_1.9.0 withr_2.5.0 xts_0.13.0
[100] scatterplot3d_0.3-43 abind_1.4-5 RCurl_1.98-1.10
[103] sp_1.6-0 nnet_7.3-18 tibble_3.2.1
[106] future.apply_1.10.0 batchelor_1.14.1 crayon_1.5.2
[109] car_3.1-1 utf8_1.2.3 RApiSerialize_0.1.2
[112] GetoptLong_1.0.5 locfit_1.5-9.7 grid_4.2.3
[115] data.table_1.14.9 vcd_1.4-11 digest_0.6.31
[118] tidyr_1.3.0 RcppParallel_5.1.7 munsell_0.5.0
[121] stringfish_0.15.7 smoother_1.1
"r.bracketedPaste": true,
"r.alwaysUseActiveTerminal": true,
"r.lsp.debug": true,
"r.rpath.linux": "R",
"r.rterm.linux": "/usr/local/bin/radian",
"r.rterm.option": [
"--r-binary=R",
"--local-history",
"--no-save",
"--no-restore"
],
"r.helpPanel.cacheIndexFiles": "Global",
"r.helpPanel.clickCodeExamples": {
"Click": "Ignore",
"Ctrl+Click": "Copy"
},
"r.plot.useHttpgd": true,
"r.rmarkdown.chunkBackgroundColor": "",
"r.rmarkdown.codeLensCommands": [
"r.runCurrentChunk",
"r.goToPreviousChunk",
"r.goToNextChunk"
],
"r.rmarkdown.knit.defaults.knitWorkingDirectory": "workspace root",
"r.session.data.rowLimit": 20000,
"r.session.levelOfObjectDetail": "Normal",
// "r.session.objectLengthLimit": 1000,
"r.session.viewers.viewColumn": {
"plot": "Active",
"browser": "Active",
"viewer": "Active",
"view": "Active",
"helpPanel": "Active"
}
Environment (please complete the following information):
- OS: Linux
- VSCode Version: 1.76.2
- R Version: 4.2.3
- vscode-R version: 2.7.2
I have the same issue. I end up with a lot of "radian" process still running at the end of the day.
For info, here is what is happening.
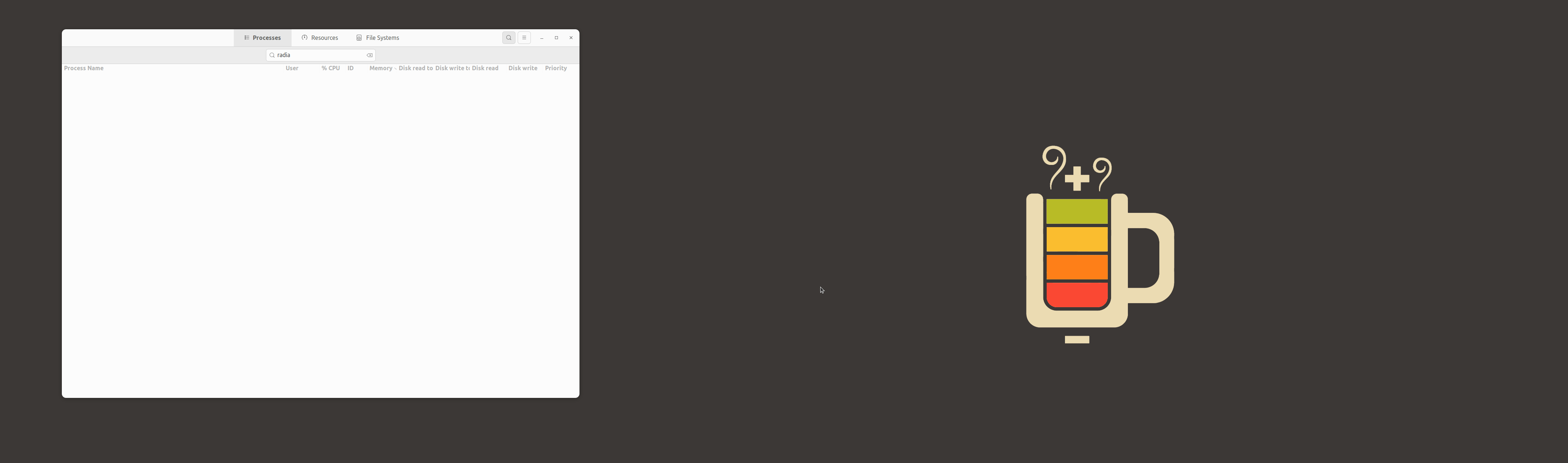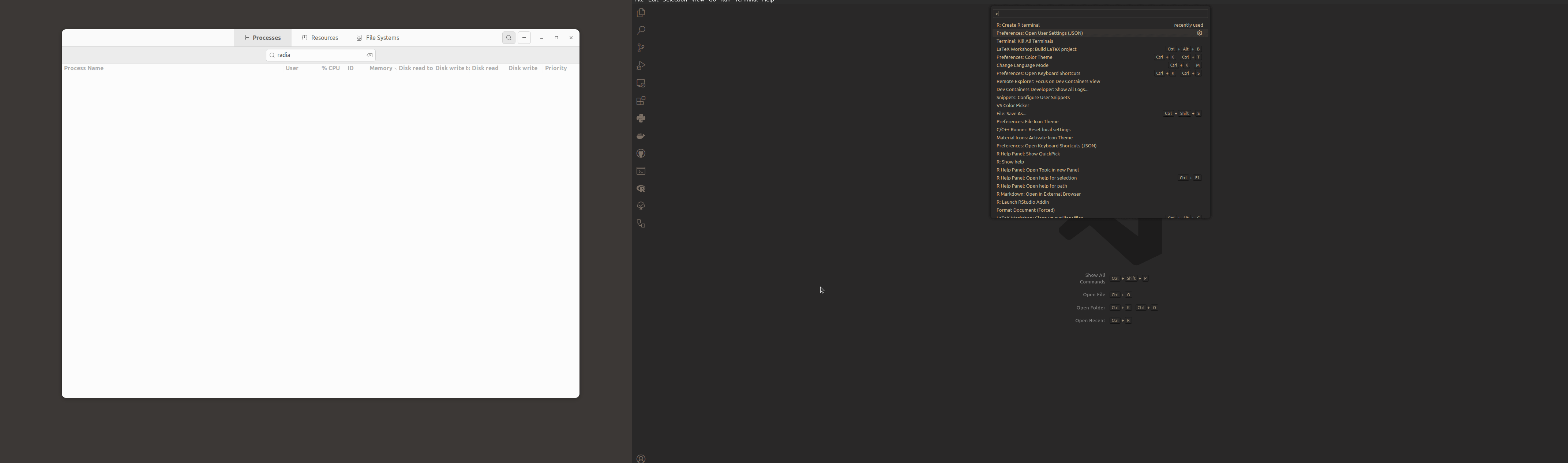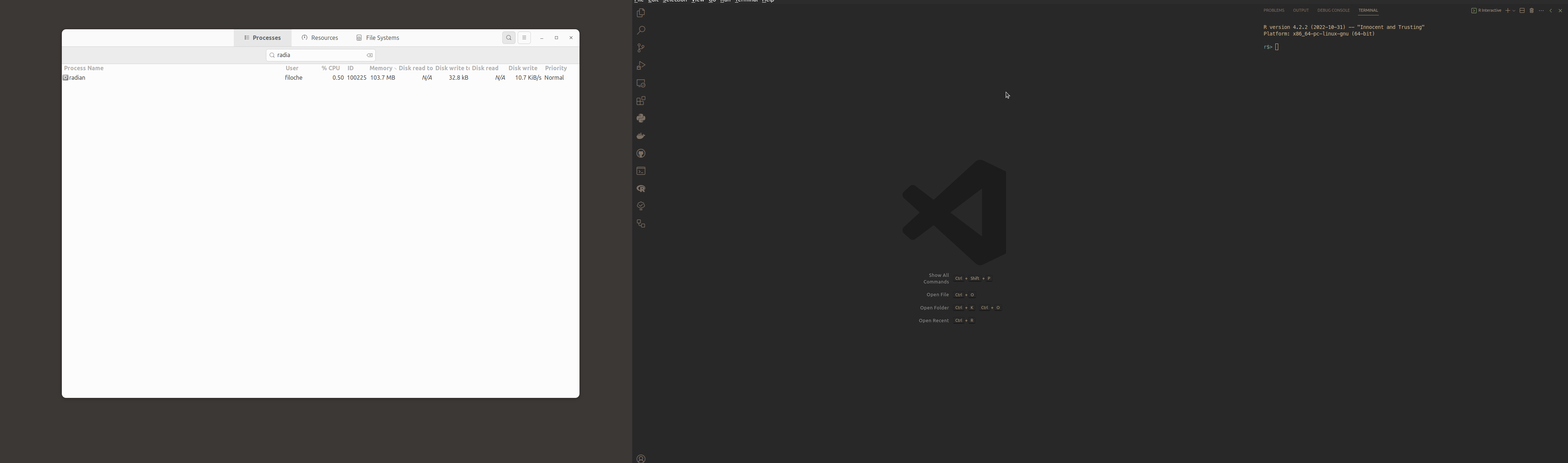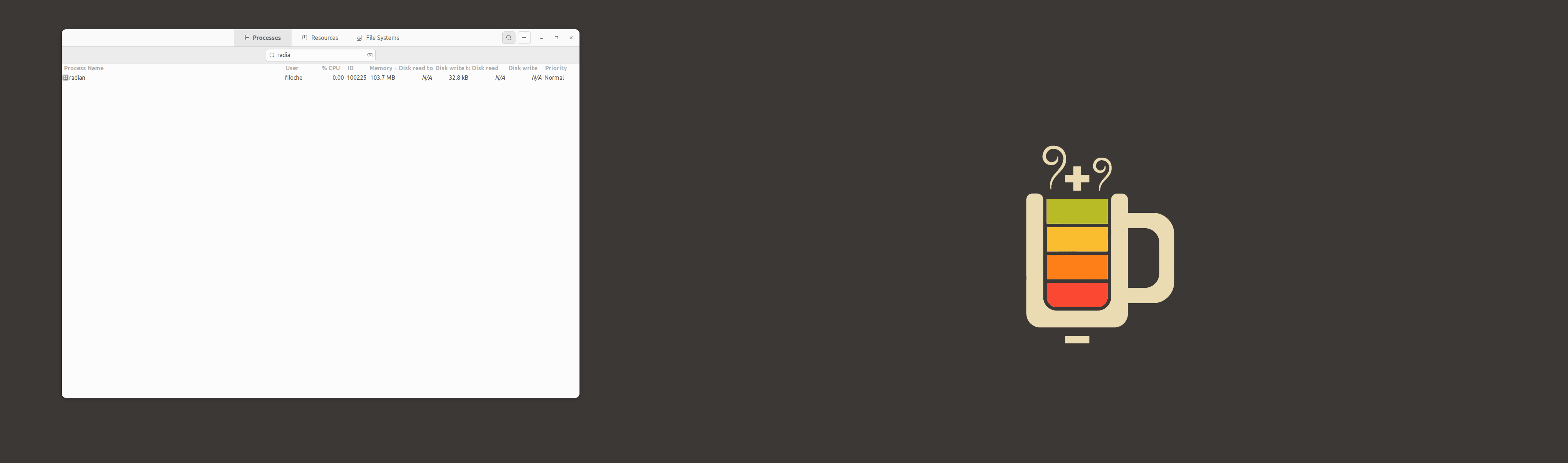
I see this every time I close VS Code on MacOS (13.3.1, M1). The R/radian process is killed if I close the (internal) terminal it's running in, but not if I close VS code altogether.
This issue is stale because it has been open for 365 days with no activity.
This issue was closed because it has been inactive for 14 days since being marked as stale.