Column Pooler - WIP
Also see issue #94
Scores 93% on MNIST dataset. Almost nothing else works.
Task List:
- [x] Merge new MNIST test runner from master
- [x] Distal Dendrites via Temporal Memory
- [ ] Column Pooler activation function
- [x] Stability & Fatigue, Multiple proximal segments
- [x] Alter learning rules
Minor Tasks:
- [ ] General cleanup & commenting
- [ ] Serialization
- [ ] Split into header & source files
- [x] Move Topology to its own file
- [ ] Make & use
InitialPermanence_t - [ ] Use camelCase everywhere:
- [ ]
stability_rate,fatigue_rate,X_act,X_inact - [ ] More descriptive names for
X_[in]act
- [ ]
- [ ] Profile & optimize
Examples:
- MNIST - Demonstrates proximal dendrites
- Columns Paper - Demonstrates distal dendrites & competition, stability
- Grid Cells - Demonstrates stability, fatigue, altered learning rules. Experiment with distal connections causing grids to align and tessellate.
Hey @ctrl-z-9000-times , this is great! A few high-level questions:
- how much is this similar to Numenta's python ColumnPooler from htmresearch?
- is the (near) goal to act as a CP, or as well to replace SP? (I know how it scores better at MNIST than current SP)
BTW, how can you mark a "draft" PR?
BTW, how can you mark a "draft" PR?
When opening a PR on github: the button has a little drop down menu inside of it, which switches between ready & draft.
- how much is this similar to Numenta's python ColumnPooler from htmresearch?
Code & API-wise, significantly different. Algorithm is the same (or will be when done), except where I've improved it.
- is the (near) goal to act as a CP, or as well to replace SP?
Both. A minimal CP contains an SP.
, except where I've improved it.
depends how you develop this. But if you start from the reference implementation, it might be valuable to have both the "original" CP, and then your improved. No problem if not, just sayin'..
I will put together a NetworkAPI Region for this.... CPRegion.
I will put together a NetworkAPI Region
Thanks, but you should hold off until the API is stable. I want to rewrite the constructor to use a structure to pass the parameters.
I rewrote the Column Pooler to use a parameter structure. Commit 7ab7ffe has 83 fewer lines of code, all of them were repeated parameter declarations.
I rewrote the Column Pooler to use a parameter structure. Commit 7ab7ffe has 83 fewer lines of code, all of them were repeated parameter declarations.
I really like that change. @dkeeney has used it in some class too. This is what I was proposing with the JSON constructor parameters (or struct), makes so many things easier..serialization, swarming. We should consider using this pattern more :+1:
I'm going to merge branch grid-cell-experiment (PR #352) into this PR and then abandon that one. They have the same code base; the only difference is the grid-cell demonstration code, which I will merge into here too (it can be removed before merging this PR into master).
I'm going to merge branch grid-cell-experiment
I'd say first merge plain CP, and then focus on grids. But ok if that way is better for you
Ive begun moving the distal dendrites code from the TM into the this class. As part of this effort i am looking for common functions which should be shared.
I intend to move the function "removeMinPermanenceSynapses" from the TM (where it is private) to the connections class where it will be a public method. I will submit a PR when i get around to it.
Ive begun moving the distal dendrites code from the TM into the this class. As part of this effort i am looking for common functions which should be shared.
so that TM would stay as is, and CP will be with all the news? :+1:
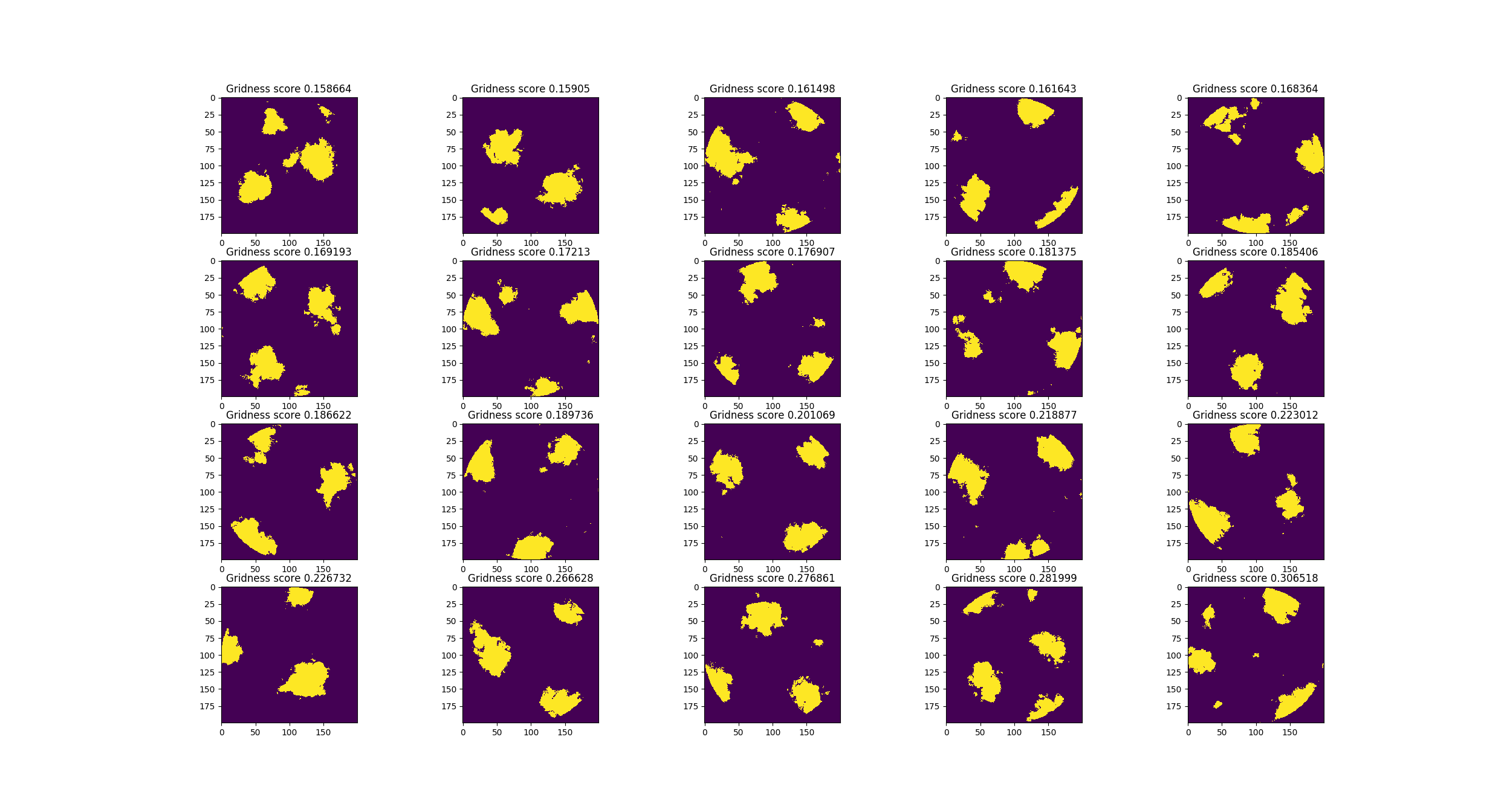
I optimized the grid cell experiment. Previously I found good parameters by hand, this time I used swarming and the AE program. Here are the improved results:
These are the best grid cells, by gridness score:
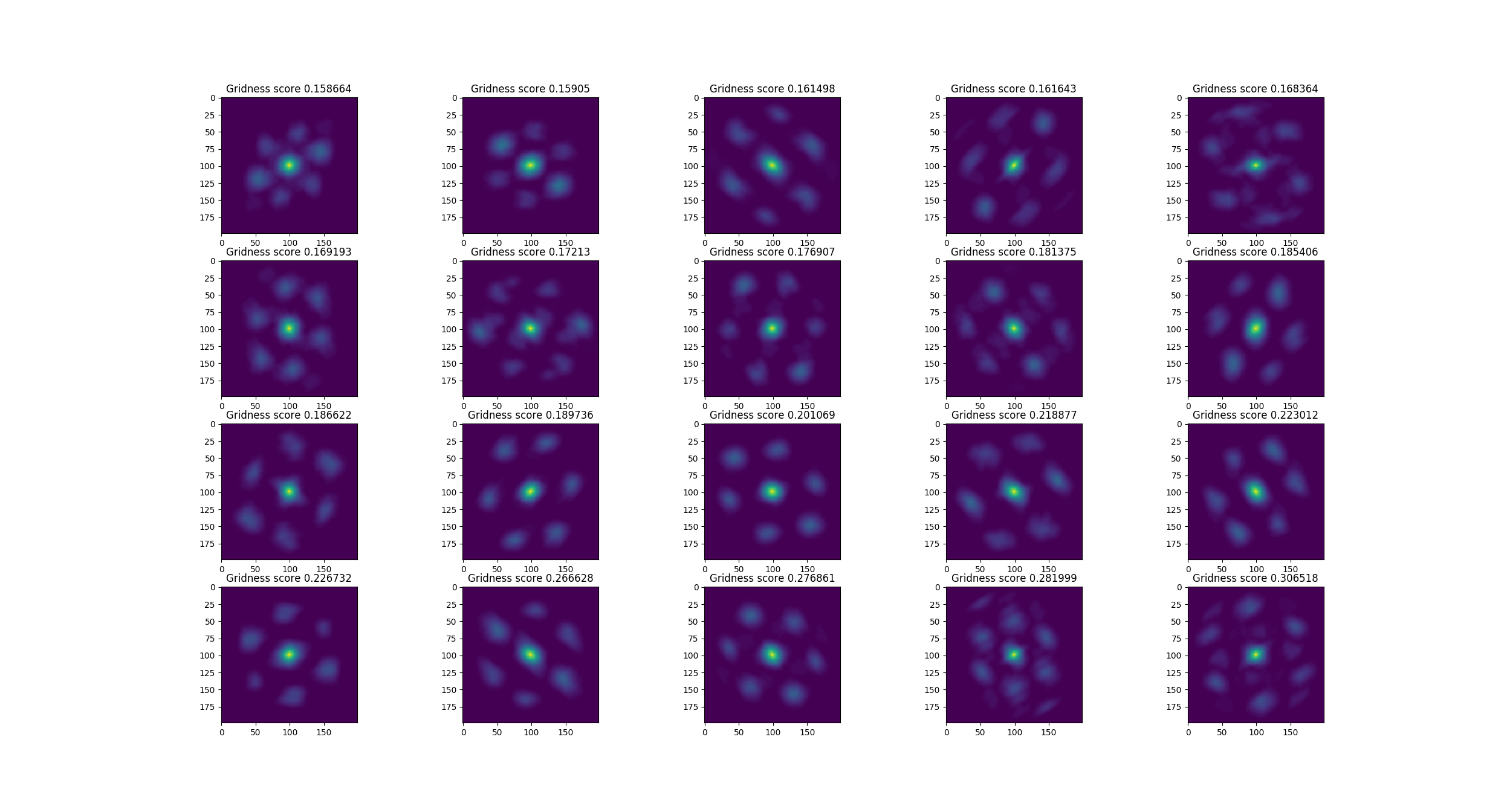
Auto-correlations of the cells shown above:
Histogram of scores for all of the cells:
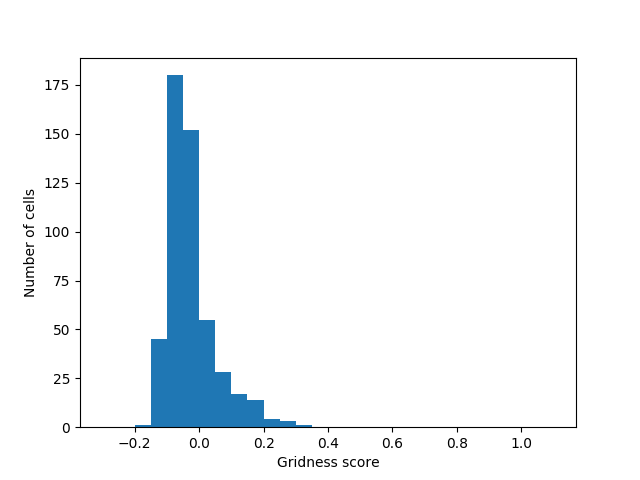
This does not perform nearly as well as the original (Kropff & Treves '08) model but I think it performs well enough to validate this model. Now I'd like to work on the distal dendrites. In theory: the distal dendrites should cause grid cells to align and tessellate. Kropff & Treves could not make their grid cells align without resorting to non-biological methods (they create the synaptic connections which they think should exist as a result of hebbian learning, but do not actually use hebbian learning to create them). I hope to apply methods from HTM-theory to the (Kropff & Treves '08) model to make grid cells align under biological constraints.