datatable
 datatable copied to clipboard
datatable copied to clipboard
missing values not handled using na_strings.
import pandas as pd
import datatable as dt
missing_values = ['', '?', 'None', 'nan', 'NA', 'N/A', 'unknown', 'inf', '-inf', '1.7976931348623157e+308', '-1.7976931348623157e+308']
train_data = "tests/smalldata/synthetic/bigIntFloatsOverflows.csv.zip"
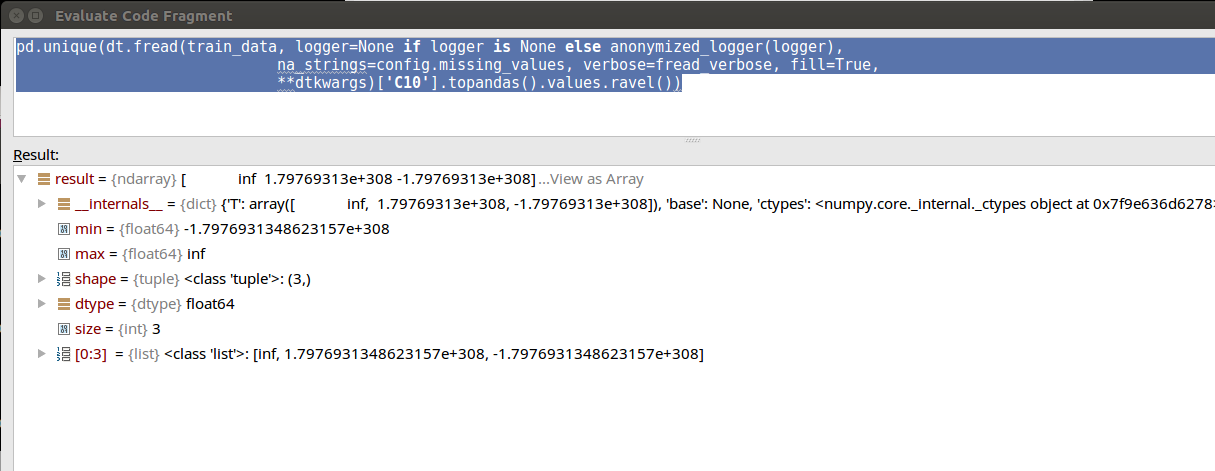
un = pd.unique(dt.fread(train_data, na_strings=missing_values, verbose=True, fill=True,)['C10'].topandas().values.ravel())
print(un)
This shows the same exact value shown in the missing_values list of strings. This used to work.
jon@pseudotensor:~/h2oai$ python dtmissingtest.py
[1] Prepare for reading
Input is assumed to be a file name.
Extracting tests/smalldata/synthetic/bigIntFloatsOverflows.csv.zip to temporary directory /tmp/tmpkj4np7g0
Using default 16 threads
fill=True (incomplete lines will be padded with NAs)
sep = <auto-detect>
Decimal separator = '.'
Quote char = (")
show_progress = True
na_strings = ["", "?", "None", "nan", "NA", "N/A", "unknown", "inf", "-inf", "1.7976931348623157e+308", "-1.7976931348623157e+308"]
+ some na strings look like numbers
+ empty string is considered an NA
strip_whitespace = True
skip_blank_lines = False
File "/tmp/tmpkj4np7g0/bigIntFloatsOverflows.csv" opened, size: 7914190
==== file sample ====
"C1","C10"
7164879580655050506,"inf"
-4848793185441839069,"inf"
2927783445302377947,"inf"
2511400862484222554,"inf"
=====================
LF character (\n) found in input, \r-only newlines will not be recognized
[2] Detect separator, quoting rule, and ncolumns
sep=',' with 100 lines of 2 fields using quote rule 0
Detected 2 columns
sep = ','
Quote rule = 0
[3] Detect column types and header
Number of sampling jump points = 101 because the first chunk was 3014.9 times smaller than the entire file
Type codes (jump 000): IF
`header` determined to be True due to column 1 containing a string on row 1 and type Int64 in the rest of the sample.
Type codes (final): IF
=====
Sampled 100 rows (handled \n inside quoted fields) at 101 jump point(s)
Bytes from first data row to the end of last row: 7914190
Line length: mean=26.40 sd=0.64 min=25 max=27
Estimated number of rows: 7914190 / 26.40 = 299780
Initial alloc = 329758 rows (299780 + 10%) using bytes/max(mean-2*sd,min) clamped between [1.1*estn, 2.0*estn]
[4] Assign column names
[5] Apply user overrides on column types
Allocating 2 column slots with 329758 rows
[6] Read the data
The input will be read in 128 chunks of size 61829 each
[7] Finalize the datatable
=============================
Read 300,009 rows x 2 columns from 7.548MB input in 00:00.022s
= 0.000s ( 1%) memory-mapping input line
+ 0.000s ( 1%) detecting parse parameters
+ 0.000s ( 2%) detecting column types using 100 sample rows
+ 0.000s ( 0%) allocating [329,758 x 2] frame (5.032MB) of which 300,009 (91%) rows used
+ 0.021s (96%) reading data using 16 threads
= 0.007s (30%) reading into row-major buffers
+ 0.000s ( 2%) saving into the output frame
+ 0.014s (64%) waiting
+ 0.000s ( 0%) creating the final Frame
Removing temporary file /tmp/tmpkj4np7g0/bigIntFloatsOverflows.csv
[ inf 1.79769313e+308 -1.79769313e+308]
jon@pseudotensor:~/h2oai$
The problem here is that the values are quoted in the file, and na_strings cannot handle quoted NA values. This was discussed long time ago, and we agreed this was a bug, however a low-priority one (given that the workaround is to include the quoted versions of same strings in the list of possible na_strings).
Ah, I see what you mean.
But dt parses the quoted thing eventually as a number it seems, else it wouldn't have truncated the numerical value to 1.79769313e+308 -1.79769313e+308 when printing out the "un" result.
So it's a real bug in the mixed way in which some quoted thing is handled, no?
I'll check some more.
still issue.