tapas
 tapas copied to clipboard
tapas copied to clipboard
Converting TaPas pretrained model to PyTorch
Hello,
as a summer project (and to learn a lot about both Tensorflow and PyTorch), I am working on incorporating a PyTorch implementation of the TAPAS algorithm into the Transformers library by HuggingFace. Similar to other models, the API would look something like this:
import pandas as pd
from transformers import TapasTokenizer, TapasForQuestionAnswering
data = {'Actors': ["Brad Pitt", "Leonardo Di Caprio"], 'Number of movies': [87, 53]}
table = pd.DataFrame.from_dict(data)
queries = ["What is the name of the first actor?", "How many movies has he played in?"]
tokenizer = TapasTokenizer.from_pretrained("tapas-base-finetuned-sqa")
model = TapasForQuestionAnswering.from_pretrained("tapas-base-finetuned-sqa")
inputs = tokenizer(table, queries, conversational=True, reset_position_index_per_cell=True, return_tensors="pt")
outputs = model(**inputs)
logits, probs, logits_aggregation, logits_cls, span_indexes, span_logits = outputs
I'm submitting this issue to get feedback on my current approach (as well as a couple of questions related to the token type embeddings), and to know whether you consent on incorporating this into the Transformers library. If yes, then I would submit this issue to the Transformers repository and start working on a pull request (hopefully with the help of other people).
To implement a new model in the Transformers library, 3 things need to be defined (each in a corresponding Python script):
- a model configuration (
configuration_tapas.py) - a model (
modeling_tapas.py) - a tokenizer (
tokenization_tapas.py)
Below, I describe my approach to each of these 3.
Configuration
As for the configuration of the base model, this seems to be a minor change of configuration_bert.py (I changed the default values of "max_position_embeddings" and "type_vocab_size"):
class TapasConfig(PretrainedConfig):
"""
model_type = "tapas"
def __init__(
self,
vocab_size=30522,
hidden_size=768,
num_hidden_layers=12,
num_attention_heads=12,
intermediate_size=3072,
hidden_act="gelu",
hidden_dropout_prob=0.1,
attention_probs_dropout_prob=0.1,
max_position_embeddings=1024,
type_vocab_size=[3, 256, 256, 2, 256, 256, 10],
initializer_range=0.02,
layer_norm_eps=1e-12,
pad_token_id=0,
gradient_checkpointing=False,
**kwargs
):
super().__init__(pad_token_id=pad_token_id, **kwargs)
self.vocab_size = vocab_size
self.hidden_size = hidden_size
self.num_hidden_layers = num_hidden_layers
self.num_attention_heads = num_attention_heads
self.hidden_act = hidden_act
self.intermediate_size = intermediate_size
self.hidden_dropout_prob = hidden_dropout_prob
self.attention_probs_dropout_prob = attention_probs_dropout_prob
self.max_position_embeddings = max_position_embeddings
self.type_vocab_size = type_vocab_size
self.initializer_range = initializer_range
self.layer_norm_eps = layer_norm_eps
self.gradient_checkpointing = gradient_checkpointing
However, the fine-tuned model has quite a lot of additional hyperparameters (as I see in tapas_classifier_model.py). I assume that these should also be defined in the class above, correct? (this is rather a question for Transformers folks)
Model
As I am not planning to retrain any pretrained/fine-tuned model, I would define the model architecture in PyTorch in modeling_tapas.py (which is very similar to modeling_bert.py - at least for the base model) and then load the weights from the Tensorflow checkpoints (for now, just MASKLM base and the fine-tuned model on SQA). The Transformers library already includes a script to convert Tensorflow checkpoints into PyTorch models, with a tutorial. This script would then be adapted for TAPAS specifically and saved as convert_tapas_original_tf_checkpoint_to_pytorch.py.
Starting off with the pretrained model (MASKLM base), this is almost identical to a pretrained BERT-base, with the only difference being the additional token_type_embeddings, as can be seen when printing out the variables of the checkpoint:
(...)
('bert/embeddings/token_type_embeddings_0', [3, 768]),
('bert/embeddings/token_type_embeddings_1', [256, 768]),
('bert/embeddings/token_type_embeddings_2', [256, 768]),
('bert/embeddings/token_type_embeddings_3', [2, 768]),
('bert/embeddings/token_type_embeddings_4', [256, 768]),
('bert/embeddings/token_type_embeddings_5', [256, 768]),
('bert/embeddings/token_type_embeddings_6', [10, 768]),
(...)
It's not really clear to me what these token type embeddings are (and in what order): token_type_embeddings_3 seem to correspond with segment ids, token_type_embeddings_1 and 2 with column and row ids, but then there are still the rank and previous answer ids. And also, what are the 2 additional token type embeddings, which do not seem to be mentioned in the paper (there are 7 token type embeddings here, but only 5 additional ids defined in the paper)? Update: appears to be "segment_ids", "column_ids", "row_ids", "prev_label_ids", "column_ranks", "inv_column_ranks" and "numeric_relations".
The core idea of loading Tensorflow weights into PyTorch is to convert variable scopes to corresponding PyTorch modules with the same name (as explained in the tutorial linked above). The code below is what I adapted from the BertEmbeddings class in the modeling_bert.py file:
class TapasEmbeddings(nn.Module):
"""Construct the embeddings from word, position and token_type embeddings.
"""
def __init__(self, config):
super().__init__()
self.word_embeddings = nn.Embedding(config.vocab_size, config.hidden_size, padding_idx=config.pad_token_id)
self.position_embeddings = nn.Embedding(config.max_position_embeddings, config.hidden_size)
self.token_type_embeddings_0 = nn.Embedding(config.type_vocab_size[0], config.hidden_size)
self.token_type_embeddings_1 = nn.Embedding(config.type_vocab_size[1], config.hidden_size)
self.token_type_embeddings_2 = nn.Embedding(config.type_vocab_size[2], config.hidden_size)
self.token_type_embeddings_3 = nn.Embedding(config.type_vocab_size[3], config.hidden_size)
self.token_type_embeddings_4 = nn.Embedding(config.type_vocab_size[4], config.hidden_size)
self.token_type_embeddings_5 = nn.Embedding(config.type_vocab_size[5], config.hidden_size)
self.token_type_embeddings_6 = nn.Embedding(config.type_vocab_size[6], config.hidden_size)
self.number_of_token_type_embeddings = len(config.type_vocab_size)
# self.LayerNorm is not snake-cased to stick with TensorFlow model variable name and be able to load
# any TensorFlow checkpoint file
self.LayerNorm = BertLayerNorm(config.hidden_size, eps=config.layer_norm_eps)
self.dropout = nn.Dropout(config.hidden_dropout_prob)
def forward(self, input_ids=None, token_type_ids=None, position_ids=None, inputs_embeds=None):
if input_ids is not None:
input_shape = input_ids.size()
else:
input_shape = inputs_embeds.size()[:-1]
seq_length = input_shape[1]
device = input_ids.device if input_ids is not None else inputs_embeds.device
if position_ids is None:
position_ids = torch.arange(seq_length, dtype=torch.long, device=device)
position_ids = position_ids.unsqueeze(0).expand(input_shape)
if token_type_ids is None:
token_type_ids = torch.zeros((*input_shape, self.number_of_token_type_embeddings), dtype=torch.long, device=device)
if inputs_embeds is None:
inputs_embeds = self.word_embeddings(input_ids)
position_embeddings = self.position_embeddings(position_ids)
token_type_embeddings_0 = self.token_type_embeddings_0(token_type_ids[:,:,0])
token_type_embeddings_1 = self.token_type_embeddings_1(token_type_ids[:,:,1])
token_type_embeddings_2 = self.token_type_embeddings_2(token_type_ids[:,:,2])
token_type_embeddings_3 = self.token_type_embeddings_3(token_type_ids[:,:,3])
token_type_embeddings_4 = self.token_type_embeddings_4(token_type_ids[:,:,4])
token_type_embeddings_5 = self.token_type_embeddings_5(token_type_ids[:,:,5])
token_type_embeddings_6 = self.token_type_embeddings_6(token_type_ids[:,:,6])
embeddings = inputs_embeds + position_embeddings + token_type_embeddings_0 + token_type_embeddings_1 + token_type_embeddings_2 + token_type_embeddings_3 + token_type_embeddings_4 + token_type_embeddings_5 + token_type_embeddings_6
embeddings = self.LayerNorm(embeddings)
embeddings = self.dropout(embeddings)
return embeddings
Alternatively, one could also let TapasEmbeddings inherit from BertEmbeddingsand overwrite the changes (I see that this was done in modeling_roberta.py for example). In the original BERT, the token type ids are a Tensor of shape (batch_size, sequence_length) in a single forward pass. However, since we now have several token type ids for every training example, I assume the shape is (batch_size, sequence_length, len(config.type_vocab_size))?
Similarly, the forward pass of BertModel (also defined in modeling_bert.py) should be updated to account for the different shape of token type ids. The best way would be to define a class called TapasModel that inherits from BertModel and overwrite the changes:
class TapasModel(BertModel):
"""
This class overrides :class:`~transformers.BertModel`. Please check the
superclass for the appropriate documentation alongside usage examples.
"""
config_class = TapasConfig
base_model_prefix = "tapas"
def __init__(self, config):
super().__init__(config)
self.embeddings = TapasEmbeddings(config)
self.init_weights()
def forward(...):
(...)
if input_ids is not None and inputs_embeds is not None:
raise ValueError("You cannot specify both input_ids and inputs_embeds at the same time")
elif input_ids is not None:
input_shape = input_ids.size()
elif inputs_embeds is not None:
input_shape = inputs_embeds.size()[:-1]
else:
raise ValueError("You have to specify either input_ids or inputs_embeds")
device = input_ids.device if input_ids is not None else inputs_embeds.device
if attention_mask is None:
attention_mask = torch.ones(input_shape, device=device)
if token_type_ids is None:
token_type_ids = torch.zeros((*input_shape, len(self.config.type_vocab_size)), dtype=torch.long, device=device)
(...)
So far, these seem to be the only differences that need to defined for the base model. However, for the fine-tuned model, the classification heads and the calculation of the loss need to be defined. To implement this in the same way as other models in the Transformers library, one could define a model TapasForQuestionAnswering which has the additional heads on top of the pretrained TapasModel. In order to later load the weights from the Tensorflow checkpoint, the names of the PyTorch modules should have the same name. It would look something like this (also to be defined in modeling_tapas.py):
class TapasForQuestionAnswering(BertPreTrainedModel):
config_class = TapasConfig
base_model_prefix = "tapas"
def __init__(self, config):
super().__init__(config)
self.tapas = TapasModel(config)
self.output = nn.Linear(config.hidden_size, config.hidden_size)
self.output_cls = nn.Linear(config.hidden_size, config.num_classification_labels)
self.output_agg = nn.Linear(config.hidden_size, config.num_aggregation_labels)
self.column_output = nn.Linear(config.hidden_size, config.hidden_size)
self.init_weights()
def forward(
self,
input_ids=None,
attention_mask=None,
token_type_ids=None,
position_ids=None,
head_mask=None,
inputs_embeds=None,
start_positions=None,
end_positions=None,
output_attentions=None,
output_hidden_states=None,
):
outputs = self.tapas(
input_ids,
attention_mask=attention_mask,
token_type_ids=token_type_ids,
position_ids=position_ids,
head_mask=head_mask,
inputs_embeds=inputs_embeds,
output_attentions=output_attentions,
output_hidden_states=output_hidden_states,
)
last_hidden_state = outputs[0] # last_hidden_state has shape (batch_size, seq_length, hidden_size)
token_logits = self.output(last_hidden_state) / config.temperature
(...)
(code from tapas_classifier_model.py and tapas_classifier_model_utils.py)
(...)
logits_aggregation = self.output_agg(last_hidden_state[:,0,:])
logits_cls = self.output_cls(last_hidden_state[:,0,:])
(...)
return outputs # (loss), logits, probs, logits_aggregation, logits_cls, span_indexes, span_logits, (hidden_states), (attentions)
Here, the code from tapas_classifier_model.py and tapas_classifier_model_utils.py should be incorporated (and translated into PyTorch code). Looking at the code, it seems that the following heads on top of the pretrained model have to be defined:
-
output_weightsandoutput_bias. These are used to compute the token logits. -
output_weights_clsandoutput_bias_cls. These are used to compute the classification logits. -
output_weights_aggandoutput_bias_agg. These are used to compute the aggregation logits. -
column_output_weightsandcolumn_output_bias. These are used to compute the column logits. These are defined in tapas_classifier_model_utils.py.
When I looked at the variables of the checkpoint of the model fine-tuned on SQA, I got this:
(...)
('bert/pooler/dense/bias', [768]),
('bert/pooler/dense/kernel', [768, 768]),
('column_output_bias', []),
('column_output_bias/adam_m', []),
('column_output_bias/adam_v', []),
('column_output_weights', [768]),
('column_output_weights/adam_m', [768]),
('column_output_weights/adam_v', [768]),
('global_step', []),
('output_bias', []),
('output_bias/adam_m', []),
('output_bias/adam_v', []),
('output_weights', [768]),
('output_weights/adam_m', [768]),
('output_weights/adam_v', [768])]
There is no aggregation/classification layer included here, probably since SQA does not require aggregation and classification.
Tokenizer
The tokenizer prepares the inputs to feed them into the model (i.e. turn a table and its queries into input ids, token type ids, etc.). This also includes truncating a table if it's too big. Here, the code of tf_example_utils.py should be incorporated (and translated into PyTorch code). The end result should be in a script called tokenization_tapas.py.
An idea could be that the tokenizer accepts Pandas dataframes as table formats, and corresponding queries as a list of strings. This will also be far from trivial, maybe one should first convert a Pandas dataframe (together with the answer coordinates and answer text) into the intermediate Interaction format, and then convert that into PyTorch Tensors.
I know that all of this is quite challenging, but it would be awesome if it would be included in the HuggingFace repo 😄
Your feedback on all of this is greatly appreciated.
Hi Niels, thanks for looking into this! I think this can be very useful to many people out there!
I didn't read your post in great detail and I am not very familiar with pytorch or the transformer library but happy to help if you have concrete questions about the TAPAS implementation.
On a higher-level, TAPAS is actually just a normal BERT model with some special embeddings to encode the table structure.
I would recommend starting to build a model that can represent a pre-trained TAPAS checkpoint. This way, you should be able to reuse most of an existing BERT implementation and you just need to add the special embeddings.
I am interested in TAPAS algorithm on pytorch. When do you plan to make it public. Thanks.
@thomasmueller-google I managed to convert tfexamples into PyTorch tensors (so I'm able to provide data to PyTorch models).
Now the next steps are defining the model configuration and the model architecture in PyTorch. It would be greatly appreciated if you had a look at my current implementation here:
-
configuration_tapas.py: this configuration class inherits from BertConfig, which in turn inherits from PretrainedConfig.- The hyperparameters are taken from
tapas_classifier_model.py. Personally, I would omit thedisabled_featuresanddisable_position_embeddingsparameters, since best performance is obtained when all features are used. Also, thedo_model_aggregationparameter can be omitted because we already have thenum_aggregation_labelsparameter, from which the former can be inferred. Correct? - Question: is it correct to state that one always has to choose one of the 4 tasks when fine-tuning? Tapas does not allow to define a task yourself, and set the hyperparameters accordingly?
- The hyperparameters are taken from
-
modeling_tapas.py: currently, I implemented the base model, which just has a number of token type embedding layers rather than just 1. The model inherits from BertModel, which in turn inherits from PretrainedModel.- Question 1: in terms of the forward pass, is it correct to state that the
token_type_idsis a Tensor of shape(batch_size, sequence_length, len(config.type_vocab_size)(assuming no features are disabled) in each forward pass? - Question 2: does the following order of features:
"segment_ids", "column_ids", "row_ids", "prev_label_ids", "column_ranks", "inv_column_ranks", "numeric_relations"correspond to the order of token_type_embeddings as found in the checkpoint:
- Question 1: in terms of the forward pass, is it correct to state that the
('bert/embeddings/token_type_embeddings_0', [3, 768]),
('bert/embeddings/token_type_embeddings_1', [256, 768]),
('bert/embeddings/token_type_embeddings_2', [256, 768]),
('bert/embeddings/token_type_embeddings_3', [2, 768]),
('bert/embeddings/token_type_embeddings_4', [256, 768]),
('bert/embeddings/token_type_embeddings_5', [256, 768]),
('bert/embeddings/token_type_embeddings_6', [10, 768])
? If yes, then I can already convert the Tensorflow checkpoint of the base model to PyTorch.
Thank you for your help and time.
@lairikeqiA as you can see this is a work in progress 😄
(I didn't have time to look at the code, I am just answering the questions)
I would also omit disabled_features and disable_position_embeddings.
Yes, you don't need do_model_aggregation it's a function of the number of labels.
run_task_main.py doesn't allow you to run with arbitrary parameters but tapas_classifier_experiment.py does (it's less convenient, though). TapasClassifierConfig is the object that stores all the options, I guess you would want to have something similar.
The shape for token_type_ids looks correct to me.
The order is defined here: https://github.com/google-research/tapas/blob/master/tapas/models/bert/table_bert.py#L36
Thank you for your response.
The base pretrained model (masklm) is converted into PyTorch. Now, I want to check whether this conversion is correct by comparing its outputs to the Tensorflow version on the same (realistic) inputs.
What is the easiest way to see the hidden states of the pretrained Tapas Tensorflow model on a particular example (table + query) (and possibly, at several locations along the depth of the model i.e. not only the final hidden states)? Also, dropout and all other nondeterministic parts should be deactivated to ensure maximal compatibility.
Hi @NielsRogge, I have converted the SQA base TF1.X model to the TF2 model. You can check the code and model in this repo. You can use the notebook for testing the inference. Currently, it doesn't contain the weight converter script. No support for training, Only inference.
Hello @NielsRogge if you want to add more information to the model output you can do so in the predictions map in tapas_classifier_model.py for example by adding model.pooled_output(), model.sequence_output() or model.get_all_encoder_layers() which hold the [CLS] token output in the last layer, the output of all the tokens in the last layer, or in all the layers, respectively. I think the pooled output should be enough.
Let me know if this makes sense. In predict mode (estimator.predict) the dropout will be disabled, so no need to worry about that. You can double check by calling it multiple times.
Ok, actually for now I just wanted to test the base model (without any heads on top), so what I did was initialize my base PyTorch model (TapasModel) with the weights of SQA base. Then, I ran the official SQA notebook, with (1) the minor change you proposed (adding model.get_pooled_output() in the predictions map), and (2) letting run_task_main.py print the pooled output of the model.
The max absolute difference turns out to be marginal (0.004), so I assume I can start working on implementing the classification heads.
Thank you! Keeping this thread open for further questions.
@thomasmueller-google @eisenjulian happy to say I've got already a working Colab that returns aggregation and classification logits of my PyTorch implementation of Tapas (you can run it from top to bottom): https://colab.research.google.com/drive/1avICEeZhQ-AH9YU2LWB3l53A1z6nLDg4?usp=sharing
To check whether the aggregation logits are exactly the same, you can compare them to the Tensorflow implementation here: https://colab.research.google.com/drive/18pWrzXi8eYwdL6xdRuhgyt5RZYwF1ej7?usp=sharing
I still have to implement 3 functions: _calculate_expected_result, _calculate_regression_loss and _single_column_cell_selection_loss. What is the best way to verify whether their implementation is correct? Should I define a separate modeling_tapas_test.py file with unit tests on a given batch? Can I print out the losses of the Tensorflow implementation without training the model?
Btw, I've already implemented the PyTorch equivalent of segmented_tensor.py here.
Wow! This is great progress! Well done!
What we do in some unit test is build an estimator and call it on a small number of inputs. I guess you could call train() or eval() to get the loss.
Here is an example where we get the eval metrics:
https://github.com/google-research/tapas/blob/master/tapas/models/tapas_classifier_model_test.py#L180
Is it possible to call estimator.evaluate() in run_task_main.py? This would be ideal, since I can then test it on a real batch from WTQ or SQA.
I tried this by adding the following in run_task_main.py:
params = dict(
batch_size=4
)
eval_input_fn = functools.partial(
tapas_classifier_model.input_fn,
name='evaluate',
file_patterns=example_file,
data_format='tfrecord',
compression_type=FLAGS.compression_type,
is_training=False,
max_seq_length=FLAGS.max_seq_length,
max_predictions_per_seq=_MAX_PREDICTIONS_PER_SEQ,
add_aggregation_function_id=do_model_aggregation,
add_classification_labels=False,
add_answer=use_answer_as_supervision,
include_id=False,
params=params
)
eval_metrics = estimator.evaluate(eval_input_fn)
I'm not that familiar with the tf.estimator API, I am using the same input_fn as for prediction (I see that this is also done in tapas_classifier_model_test.py). However, I got the following error:
ValueError: input_fn (functools.partial(<function input_fn at 0x7fe7da8aeb70>, name='evaluate', file_patterns='/content/drive/My Drive/Tapas/WTQ/results/wtq/tf_examples/random-split-1-dev.tfrecord', data_format='tfrecord', compression_type='--init_checkpoint=tapas_wtq_wikisql_sqa_masklm_base/model.ckpt', is_training=False, max_seq_length=512, max_predictions_per_seq=20, add_aggregation_function_id=True, add_classification_labels=False, add_answer=True, include_id=False, params={'batch_size': 4})) does not include params argument, required by TPUEstimator to pass batch size as params["batch_size"]
It says that the input_fn does not include the params argument, whereas it clearly does?
notebook to reproduce: https://colab.research.google.com/drive/18pWrzXi8eYwdL6xdRuhgyt5RZYwF1ej7#scrollTo=wQJ5zQLOxWbs
UPDATE: solved it by removing the params argument above (as it's already included in the input_fn, with only 1 key namely the gradient_accumulation_steps) and adding the following to dataset.py in the read_dataset function:
if "batch_size" in params:
batch_size = params["batch_size"]
else:
batch_size = 4
Currently, I have a working implementation of TapasForQuestionAnswering in PyTorch, with an associated TapasTokenizer that prepares the data for the model. It removes the need for an intermediate Interaction format, and uses Pandas dataframes as the central object. You can try it out here.
However, I've got a question related to the API of the tokenizer.
Inference
When you provide TapasTokenizer with a table (Pandas dataframe) and associated queries (a list of strings), it creates the input_ids, attention_mask and token type ids. So, inference works (as can be seen in the notebook above).
Training
However, when you also want to calculate the loss (in order to train/fine-tune the model), a number of additional things need to be provided to the model:
-
label_ids- labels per token, of shape (batch_size, seq_len) -
answer- answer (float value) for every example in the batch if present, of shape (batch_size,) -
numeric_values- numeric values of every token, of shape (batch_size, seq_len) -
numeric_values_scale- scale of the numeric values of every token, of shape (batch_size, seq_len) -
aggregation_function_id(in case of aggregation) for every example in the batch - of shape (batch_size,) -
classification_class_index(in case of classification) for every example in the batch - of shape (batch_size,).
For text classification with models like BERT and RoBERTa, the API in the Transformers library is straightforward: people just need to prepare a table of text and label columns, and the tokenizer can then be used to create the input_ids, attention_mask and token_type_ids based on the text, whereas the labels should be prepared by the user in case they want to compute a loss.
However, in case of TAPAS - how should people prepare their data? My idea is to let people prepare the data in the TSV format of SQA, with the columns "position" (for conversational setups), "table_file", "answer_coordinates", "answer_text", "aggregation", "float_answer" and "class_index". Currently, TapasTokenizer can create label_ids, answer, numeric_values and numeric_values_scale based on the "table", "answer_coordinates" and "answer_text" columns. The answer, aggregation_function_id and classification_class_index tensors should still be prepared by the user.
It is clear to me how this works for SQA - see the notebook above, as the answer_coordinates and answer_texts columns are always populated (because SQA only has cell selection questions). But how does this table look like in case of WTQ, where there are scalar answers and ambiguous answers? Does the model create label_ids in that case, or are they just set to zero?
Let's take a concrete example: question 3 of figure 3 from the original paper. Are the answer_coordinates and answer_text fields populated in that case? I see here that the answer_coordinates are set to (-1, -1), but the answer texts are still created. So that means the answer coordinates are (-1, -1) and the answer text is equal to "2"? @thomasmueller-google
Hi Niels!
In the case of WTQ we have some special logic that tries to find the answer text in the table or that populates the float_value
field if the answer is a real number.
The logic is here:
https://github.com/google-research/tapas/blob/master/tapas/utils/interaction_utils_parser.py
For WTQ parse_question will be called with mode REMOVE_ALL. (The same code is used for WikiSQL where we have the supervised mode that uses the coordinates extraction from the SQL and the weakly-supervised mode where we do the same as for WTQ.)
_parse_answer_coordinates is the code that searches for the answer.
_parse_answer_float will try to parse the text as a float.
_parse_answer_coordinates uses this linear optimization code but is actually just extracting the first text match of
the answer in the table.
Hi @thomasmueller-google and @NielsRogge
I was playing around but have integrated interaction_utils_parser.py (I don't know if @NielsRogge has already completed it)
Here are the test result of the most imp function parse_questions
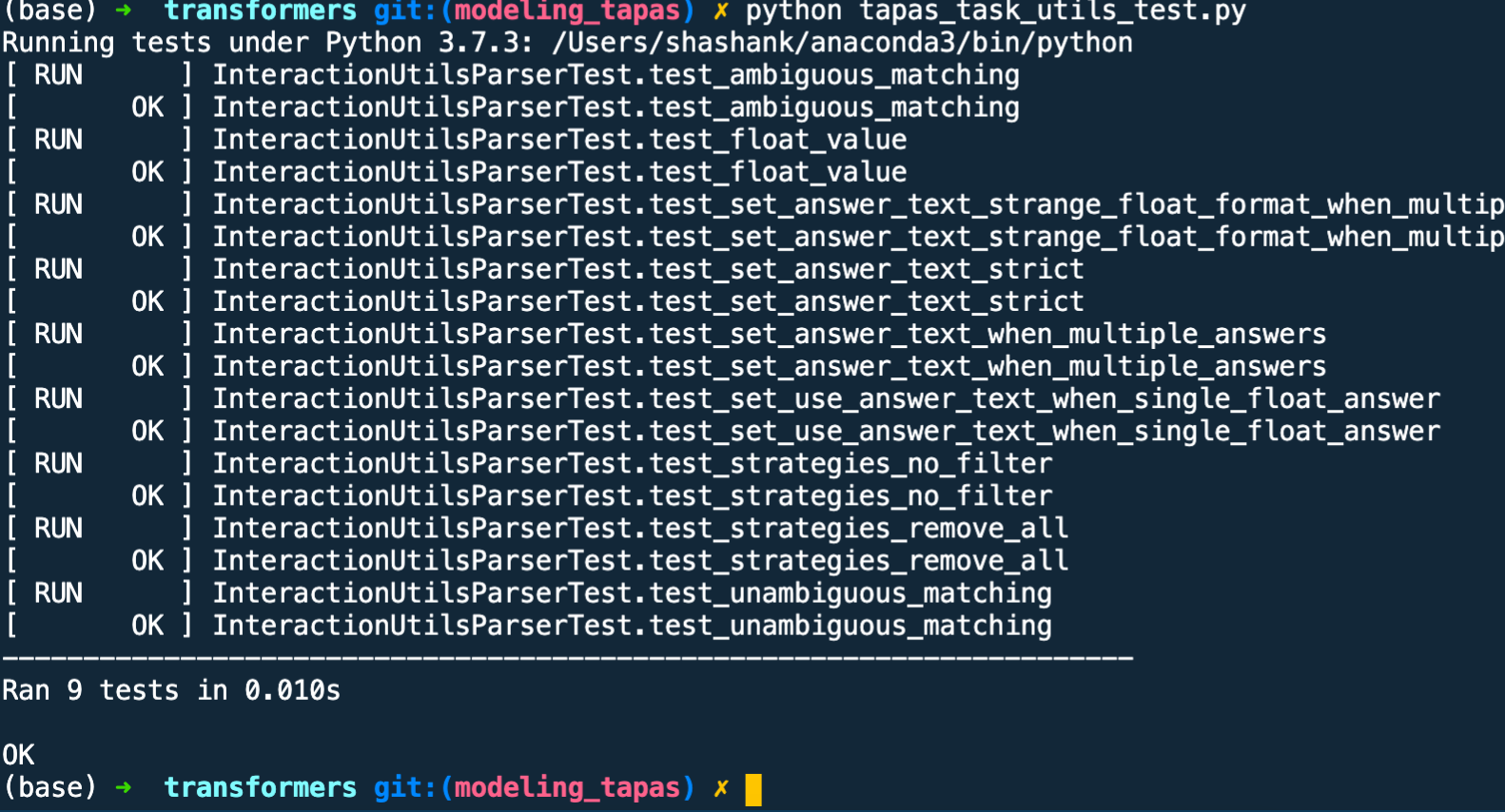
Will create a PR...
Hey @shashankMadan-designEsthetics you mean make a PR to modeling_tapas? Thank you!
I currently am working on a new branch based on the modeling_tapas branch which includes a lot of improvements, including support for Tabfact (from the follow-up paper of Tapas).
Hey @shashankMadan-designEsthetics you mean to make a PR to
modeling_tapas? Thank you!I currently am working on a new branch based on the
modeling_tapasbranch which includes a lot of improvements, including support for Tabfact (from the follow-up paper of Tapas).
Yep, @NielsRogge on the forked repo which has the branch modeling_tapas.
Let me run up on your code once again to see how to best refactor my code on it!
will look into Tabfact Sounds Interesting ;)
Thanks for a quick reply and support
Hi Niels!
In the case of WTQ we have some special logic that tries to find the answer text in the table or that populates the
float_valuefield if the answer is a real number.The logic is here:
https://github.com/google-research/tapas/blob/master/tapas/utils/interaction_utils_parser.py
For WTQ
parse_questionwill be called with modeREMOVE_ALL. (The same code is used for WikiSQL where we have the supervised mode that uses the coordinates extraction from the SQL and the weakly-supervised mode where we do the same as for WTQ.)
_parse_answer_coordinatesis the code that searches for the answer._parse_answer_floatwill try to parse the text as a float.
_parse_answer_coordinatesuses this linear optimization code but is actually just extracting the first text match of the answer in the table.
I want to convert WTQ to SQA, how should I run this script https://github.com/google-research/tapas/blob/master/tapas/utils/interaction_utils_parser.py?
I am not understanding the input format to the parse_question method.
One training example in WTQ tsv looks like this..... id annotator position question table_file answer_coordinates answer_text nt-14053 0 0 what boats were lost on may 5? table_csv/203_386.csv ['(1, 1)', '(2, 1)'] ['U-638', 'U-531']
I want to know the type of table message and question message.
Hi @NielsRogge I am preparing data for weak supervision. for some questions I have no answer for float_answer column and also I have no answer for answer_coordinates column how to handle this