Refactor images, start from same base
As discussed with @benz0li, we could use the b-data stack throughout, with more consistency and good R/Python support. First we would need to think about similarities/differences and pros/cons, leaving this as an open question, I'm a bit out of my depth, so any advice / PRs welcome : )
Feedback regarding image size (uncompressed): glcr.b-data.ch/jupyterlab/r/tidyverse (3.18 GB) vs rocker/tidyverse (2.38GB):
glcr.b-data.ch/jupyterlab/r/tidyverse is 800 MB bigger because
- code-server is bigger than RStudio Server OSE[^1]
- there is JupyterLab (incl. dependencies) installed
- some R packages have been moved up the build chain
[^1]: There are many Code extensions pre-installed.
Feedback regarding image size (uncompressed): glcr.b-data.ch/jupyterlab/python/scipy (4.35 GB) vs jupyter/scipy-notebook (4.14GB).
There are too many differences (see below) and it is sheer coincidence that the images are almost the same size.
JupyterLab R docker stack vs Rocker images (versioned stack):
Differences
- Base image: Debian instead of Ubuntu
(CUDA-enabled images are Ubuntu-based)- Unminimized, i.e. including man pages
- IDE: JupyterLab + code-server instead of RStudio
- Source builds[^1] installed for
- Python
- Git
- Git LFS
- Shell: Zsh
- Additional image
glcr.b-data.ch/jupyterlab/r/qgisprocess - Missing images:
.../geospatial:dev-osgeo.../shiny.../shiny-verse
[^1]: Newer versions than the distros's repository. Installed at /usr/local/bin.
Similarities
- Image names
Exceptions:.../vervs.../r-ver.../basevs.../rstudio
- Installed R packages
- some R packages have been moved up the build chain
JupyterLab Python docker stack vs Jupyter Docker Stacks:
Differences
- Base image: Debian instead of Ubuntu
- Unminimized, i.e. including man pages
- IDE: code-server next to JupyterLab
- Just Python – no Conda / Mamba
- GPU accelerated images available
(CUDA-enabled images are Ubuntu-based) - Source builds[^1] installed for
- Python
- Git
- Git LFS
- Shell: Zsh
- The
scipyimage includes Quarto and TinyTeX
[^1]: Newer versions than the distro's repository. Installed at /usr/local/bin.
Similarities
- Image names
- b-data's docker stack only offers
baseandscipy[^2]
- b-data's docker stack only offers
- Installed Python packages
[^2]: Any other Python package can be installed at user level.
👉 Because my images allow (bind) mounting the whole home directory, user data is persisted – i.e. survives container restarts. (Cross reference: https://github.com/jupyter/docker-stacks/issues/1478)
b-data's JupyterLab docker stack:
Pros
- All JupyterLab docker stacks include the same tool set
- Integrated Development Environments: JupyterLab + code-server
- Programming Language: Python (+ R or Julia)
- GPU accelerated images available
- Data Science Dev Containers with similar features
ℹ️ The JupyterLab docker stacks and Data Science Dev Containers also support rootless mode.
Cons
- b-data is a one-man GmbH (LLC)
- Time for image maintenance is limited
- No financial support (sponsorship)
- Execption: JupyterLab R docker stack ℹ️ Work partially funded by Agroscope.
- No testing of docker images
- Using own Docker Registry
- No plan to release on Docker Hub or Quay
Any other similarities/differences and pros/cons that come to your mind?
- @mathbunnyru: Regarding Jupyter Docker Stacks vs JupyterLab Python docker stack
- @eitsupi: Regarding Rocker images (versioned stack) vs JupyterLab R docker stack
I would like to give @Robinlovelace a solid base for his decision on how to refactor the images (possibly use b-data's image as a base).
I would also add as pros of https://github.com/jupyter/docker-stacks:
- automatic updates (including security updates of Ubuntu base image and Python packages)
- better image testing
- better image tagging
- readable build manifests for images
And cons:
- building one image set at a time (this is a choice the project made, but still, it might be a downside for people who want old python version with modern packages at the same time)
@mathbunnyru Thank you for the feedback.
I would also add as pros of https://github.com/jupyter/docker-stacks:
- automatic updates (including security updates of Ubuntu base image and Python packages)
- better image testing
- better image tagging
- readable build manifests for images
I fully agree. Cross reference regarding image tagging and readable build manifests: https://github.com/jupyter/docker-stacks/wiki/2023-11
And cons:
- building one image set at a time (this is a choice the project made, but still, it might be a downside for people who want old python version with modern packages at the same time)
Same for b-data's JupyterLab docker stacks. Rocker images (versioned stack) handles this differently.
Many thanks for all the detailed info + thoughts. At present I'm erring towards b-data's JupyterLab docker stack with data science devcontainers.
@eitsupi Any feedback from your side?
I'm sorry, but since I don't know the context, I don't think I can give any particular advice.
If I had to say, I'm very reluctant to support Python on https://github.com/rocker-org/rocker-versioned2, unlike @cboettig and @yuvipanda, and think it's better for users to install their favorite version of Python using micromamba, rye, or something. (Additionally, R can now be installed using rig, making it very easy to install your favorite R version.)
Thanks all, any other general thoughts welcome. Seeing as we're talking about Python a Python-focussed image makes sense, and good idea re. rig, do you know of any example Dockerfiles that use it @eitsupi ?
re. rig, do you know of any example Dockerfiles that use it @eitsupi ?
The rig repository has it. https://github.com/r-lib/rig/blob/1e335785f95c3669bf04a43bc6a0da4862e401db/containers/r/Dockerfile
Just install it using curl and run the rig add command.
Awesome. Many thanks!
@Robinlovelace Great discussion and tricky issues here. Love the work you are doing in this space and bridging between these communities.
For a geospatial-focused python docker images, I would definitely look to the Pangeo stack: https://github.com/pangeo-data/pangeo-docker-images. @yuvipanda can correct me if I'm wrong, but I believe these are derived from the standard Jupyter stack linked above, and importantly (imo) they include prebuilt images with gpu support which can be a common stumbling block. Note that the Microsoft Planetary Computer docker images are also derived from the Pangeo stack -- it's well maintained and widely used.
As @eitsupi mentions, @yuvipanda and I do want to see good native python support in Rocker, especially when it comes to geospatial. As you know, there really isn't a clean separation of R from python here -- the core geospatial C libraries like gdal have (technically optional but important) dependencies on python already. I also think there's a compelling case for users, instructors & platform providers to want to provide a similar base that can work across Python and R -- especially when a lot of the heavy lifting is being done by the same OSGeo C libraries, it makes sense to have access to the same versions of those libraries from both ecosystems. Lastly, R users in particular may benefit from a more 'batteries included' approach to python, rather than navigating the wide space of miniconda, conda, mamba, pyenv, system python and so forth with reticulate, especially when it comes to areas like geospatial where these packages are calling system c libraries (or coming prepackaged with binaries). It's relatively easy to get into a mess. https://github.com/rocker-org/rocker-versioned2/pull/718 looks like a promising setup, but we're still testing.
Anyway, don't mean to say that you should definitely be using rocker for this, but only that it's a use case that we're also trying to address in the rocker/geospatial images already. The pangeo stack is already solid, and I'm glad @yuvipanda has started bringing his expertise from there over to us in rocker :pray: .
For a geospatial-focused python docker images, I would definitely look to the Pangeo stack: https://github.com/pangeo-data/pangeo-docker-images. @yuvipanda can correct me if I'm wrong, but I believe these are derived from the standard Jupyter stack linked above, and importantly (imo) they include prebuilt images with gpu support which can be a common stumbling block. Note that the Microsoft Planetary Computer docker images are also derived from the Pangeo stack -- it's well maintained and widely used.
There is glcr.b-data.ch/jupyterlab/cuda/r/qgisprocess (CUDA-enabled JupyterLab R docker stack)
And there is glcr.b-data.ch/jupyterlab/cuda/qgis/base (CUDA-enabled JupyterLab QGIS docker stack):
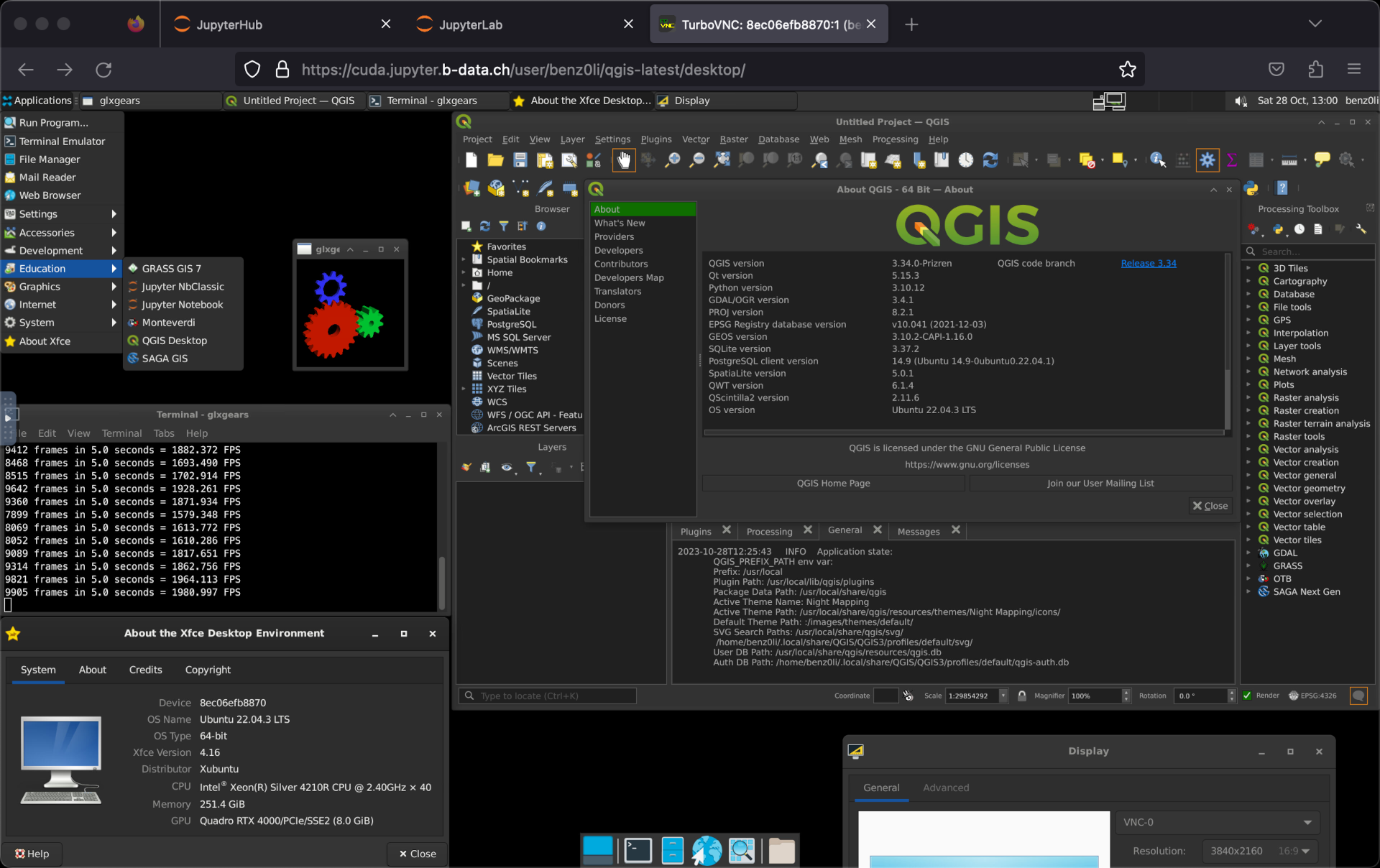
Qgis in browser with full GUI. Wow!
Qgis in browser with full GUI. Wow!
Including HiDPI support: https://github.com/b-data/jupyterlab-qgis-docker-stack/blob/main/NOTES.md#custom-display-resolution
Hi @benz0li FYI we've re-visited this, with a specific need for cross-language containers. Having looked at the images in b-data and having tried a few other options, including rocker (see https://github.com/rocker-org/rocker-versioned2/issues/860 ), we're currently looking to use pixi for a simple way to manage images with R and Python and one day Julia. Just wondering, do you have well-supported cross-language images? I've been looking at the docs here but cannot find the
docker pull ...
command that will give us a solid multi-language container with R and Python to run both books, and maybe one day a Julia book too.
@Robinlovelace this is a great question, but I think a good answer needs more specificity on what we mean by 'multi-language container'. (For instance, the examples you mentioned are all already used in multi-language settings, but that doesn't mean the serve the geocompx needs well).
For instance, for some communities, when they say "python" they mean python only via conda environments (including a lot of pangeo / python geospatial world). Others are more agnostic about this or prefer pip-based channels.
Specifically for spatial, I think that having all libraries (python and R) using a modern GDAL and complete GDAL is super important. But as you know, the majority of options out there love packaging with old versions of GDAL and with versions of GDAL that are built without support arrow/parquet drivers.
Geospatial ML stuff that needs the massive CUDA libraries for GPU-accelerated tasks is obviously another wrinkle not everyone needs.
And of course python being what it is, we can hit incompatible environments -- I have some spatial projects that wind up with a hard dependency on, say, pandas < 2.0 and others with pandas >= 2.0.
My point here is that I don't think we'll ever have a one-size-fits-all cross-language container. But a geocompx-specific container(s) seems better defined. I think we do need good base images so that such containers don't need to do everything from scratch, and maybe we're not there yet either. (especially in supporting IDE/interactive use -- I've recently followed the examples of @benz0li and others in seeing JupyterLab-based containers as the best way to support both R and python in the same interactive environment). I think we could also benefit from more streamlined ways to for users to customize and extend containers and build on each-other's extensions (e.g. @eeholmes has done some impressive work for this at NOAA) but it's still harder than it should be!
Thanks @cboettig that is really helpful. Agreed re. specificity and in fact I think we can be specific and objective: we need a container that can:
- [ ] Reproduce Geocomputation with R, with
bookdown::serve_book(".") - [ ] Reproduce Geocomputation with Python, with
quarto preview - [ ] Work in a devcontainer
- [ ] Be resource efficient
- [ ] Not require too much tinkering
It seems to me that pixi is a good way to manage cross-language environments. According to @MartinFleis, because it uses conda, will share deps where possible. Agree re. out-of-date deps but I think we don't need cutting-edge GDAL etc for our needs. Any thoughts on that v. welcome.
Working hypothesis is that extending https://github.com/geocompx/docker/blob/master/pixi-r/Dockerfile will be the quickest way to meet those criteria but not sure and feel like I'm trying things blind, very happy to start with other things and test different approaches :pray:
Just wondering, do you have well-supported cross-language images?
@Robinlovelace Yes.
Both JupyterLab Julia docker stack and JupyterLab R docker stack include Python.
- https://github.com/b-data/jupyterlab-julia-docker-stack/blob/main/VERSION_MATRIX.md
- https://github.com/b-data/jupyterlab-r-docker-stack/blob/main/VERSION_MATRIX.md
@Robinlovelace Regarding «Work in a devcontainer»:
But I think R qgisprocess is the one you are looking for: devcontainer.json, R.Dockerfile.
ℹ️ R and Julia images have Python (currently v3.11.5[^1]) installed, too.
Originally posted by @benz0li in https://github.com/geocompx/geocompy/issues/188#issuecomment-1700986248
[^1]: currently v3.12.6.
The Data Science dev containers are based on the language docker stacks.
The language docker stacks are siblings^1 of the JupyterLab docker stacks.
Good stuff, thanks @benz0li. I've had a look at https://github.com/b-data/data-science-devcontainers/blob/main/.devcontainer/R.Dockerfile and it looks great. My issue at the moment is I'm not sure how best to build on it. Any guidence welcome, starting with your recommended variant as basis of
FROM ...
first line.
@Robinlovelace Just in case it's helpful, I wanted to also share some examples from @eeholmes + co here: https://nmfs-opensci.github.io/container-images/ (github repo here: https://github.com/nmfs-opensci/container-images/, shows how to customize images, images also automatically built and served through github container registry).
Many thank Carl, these look very helpful. Will do some tests and report back.
Update here, after going down a pixi rabbit hole, it seems there are some issues with them, including image size and setting-up global or always-enabled environments. So plan A is to continue tests with mamba and rocker-based images now.
See #70, https://github.com/prefix-dev/pixi-docker/issues/42
Interestingly on the topic of image sizes, it seems to me that the image built with pip install:
https://github.com/geocompx/docker/blob/85a693cd2b6eb0a94037a87a0326d4afb0c318fa/python/Dockerfile#L13
Is smaller that the same image build with conda installation:
https://github.com/geocompx/docker/blob/85a693cd2b6eb0a94037a87a0326d4afb0c318fa/mamba-py/Dockerfile#L6
The sizes from those are:
| docker:python | docker pull ghcr.io/ghcr.io/geocompx/docker:python Python image + geo pkgs | |
and
| docker:mamba-py | docker pull ghcr.io/ghcr.io/geocompx/docker:mamba-py | |
Heads-up @martinfleis (maybe the difference will be small when R packages are added, but the above does not support the idea that conda-based installations are smaller, right?) and @benz0li, the above comparison provides :+1: for your recommended approach. Multiple rabbit holes to go down here, maybe just starting with vanilla python and R (well Rocker) images is the best way forward after all...
But you're not comparing equivalent environment, keep that in mind. There are two main differences - your environment.yml specifies more dependencies, notably quite heavy hvplot suite. Second difference is GDAL. In wheels, for example in pyogrio, we don't include all the drivers. For example, libkml is quite heavy and there might be some other that are excluded from wheels but included on conda-forge.
Also, the Python Docker already contains Python, while I believe that the micromamba container does not and Python is part of the installation.