pca
 pca copied to clipboard
pca copied to clipboard
Principal component analysis (PCA) in Ruby
Principal Component Analysis (PCA)
Principal component analysis in Ruby. Uses GSL for calculations.
PCA can be used to map data to a lower dimensional space while minimizing information loss. It's useful for data visualization, where you're limited to 2-D and 3-D plots.
For example, here's a plot of the 4-D iris flower dataset mapped to 2-D via PCA:
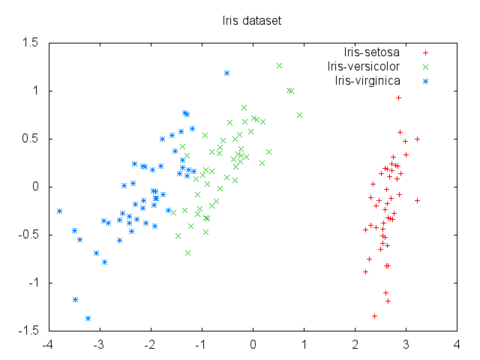
PCA is also used to compress the features of a dataset before feeding it into a machine learning algorithm, potentially speeding up training time with a minimal loss of data detail.
Install
GSL must be installed first. On OS X it can be installed via homebrew: brew install gsl
gem install pca
Example Usage
require 'pca'
pca = PCA.new components: 1
data_2d = [
[2.5, 2.4], [0.5, 0.7], [2.2, 2.9], [1.9, 2.2], [3.1, 3.0],
[2.3, 2.7], [2.0, 1.6], [1.0, 1.1], [1.5, 1.6], [1.1, 0.9]
]
data_1d = pca.fit_transform data_2d
# Transforms 2d data into 1d:
# data_1d ~= [
# [-0.8], [1.8], [-1.0], [-0.3], [-1.7],
# [-0.9], [0.1], [1.1], [0.4], [1.2]
# ]
more_data_1d = pca.transform [ [3.1, 2.9] ]
# Transforms new data into previously fitted 1d space:
# more_data_1d ~= [ [-1.6] ]
reconstructed_2d = pca.inverse_transform data_1d
# Reconstructs original data (approximate, b/c data compression):
# reconstructed_2d ~= [
# [2.4, 2.5], [0.6, 0.6], [2.5, 2.6], [2.0, 2.1], [2.9, 3.1]
# [2.4, 2.6], [1.7, 1.8], [1.0, 1.1], [1.5, 1.6], [1.0, 1.0]
# ]
evr = pca.explained_variance_ratio
# Proportion of data variance explained by each component
# Here, the first component explains 99.85% of the data variance:
# evr ~= [0.99854]
See examples for more. Also, peruse the source code (~ 100 loc.)
Options
The following options can be passed in to PCA.new:
| option | default | description |
|---|---|---|
| :components | nil | number of components to extract. If nil, will just rotate data onto first principal component |
| :scale_data | false | scales features before running PCA by dividing each feature by its standard deviation. |
Working with Returned GSL::Matrix
PCA#transform, #fit_transform, #inverse_transform and #components return instances of GSL::Matrix.
Some useful methods to work with these are the #each_row and #each_col iterators,
and the #row(i) and #col(i) accessors.
Or if you'd prefer to work with a standard Ruby Array, you can just call #to_a and get an array of row arrays.
See GSL::Matrix RDoc for more.
Plotting Results With GNUPlot
Requires GNUPlot and gnuplot gem.
require 'pca'
require 'gnuplot'
pca = PCA.new components: 2
data_2d = pca.fit_transform data
Gnuplot.open do |gp|
Gnuplot::Plot.new(gp) do |plot|
plot.title "Transformed Data"
plot.terminal "png"
plot.output "out.png"
# Use #col accessor to get separate x and y arrays
# #col returns a GSL::Vector, so be sure to call #to_a before passing to DataSet
xy = [data_2d.col(0).to_a, data_2d.col(1).to_a]
plot.data << Gnuplot::DataSet.new(xy) do |ds|
ds.title = "Points"
end
end
end
Sources and Inspirations
- A tutorial on Principal Components Analysis (PDF) a great introduction to PCA
- Principal Component Analyisis Explained Visually
- scikit-learn PCA
- Lecture video and notes (requires Coursera login) from Andrew Ng's Machine Learning Coursera class
- Implementing a Principal Component Analysis (PCA) in Python step by step
- Dimensionality Reduction: Principal Component Analysis in-depth