Append directly on/to disk
I wonder if it's possible to row append a dataset directly to a dataset stored on disk?
currenlty to append two dataframes together I need to do
a <- read.fst(file1, as.data.table=T)
b <- read.fst(file2, as.data.table=T)
write.fst(file3,rbindlist(list(a,b)))
If both a and b are large then R may run out memory. So it would prefer to have a function to append.fst e.g.
append.fst(out_file, c(infile1, infile2))
Hi @xiaodaigh, thanks for your question!
The ability to append fst files to an existing fst file is planned for release v0.9.0. That's the release after the coming CRAN release v0.8.0. The format is already prepared for 'multiple chunk' data and it will support also adding chunks with different fst versions.
In addition to that, appending columns will be also be supported and it will be possible to append rows (chunks) and columns in random order (so append a chunk and then a column and later a new chunk with more columns than the first, see also #39).
To make that possible, the fst format is basically just a collection of random-sized columnar vectors, glued together with small meta-data blocks. It doesn't matter where in the file the actual columnar data is stored, with the correct meta-data 'glue', the dataset can always be reconstructed. Each chunk or column mutation will incur a small penalty due to additional seek operations in the file, but that will only become an issue with a very large number of mutations on the file (especially with modern SSD's). To maintain data integrity, each meta-data block is hashed (and optionally also the data blocks)
Append operations will be possible with two fst files (requiring very little memory) or with a fst file and an in-memory table (requiring only memory for the in-memory table). So with that, in effect, you can 'stream' data to a fst file from any (chunked) source.
Because the actual in-file location of the columnar vector data is not important, chunks can be appended without decompressing and recompressing data. So appending compressed fst files will be even faster than appending uncompressed fst files (different compression levels will be present in the file after that however, so perhaps recompression should be a user-option)
If you have any additional questions, please let me know!
@MarcusKlik is the ability to append fst file to an existing fst file implemented in recent fst versions? I think I am a couple of step behind I think at version fst 0.7.3.
Hi @wei-wu-nyc, thanks for your question. The fst file format is already prepared to make column- and row binding possible. That means that the meta-data for current fst files has free space to add pointers to new horizontal- or vertical chunks of data (but they are not filled for current fst files).
The feature is planned for milestone fst v0.8.8, after initial data.table and dplyr interfaces are available. Those interface will be requiring row- and column binding for more advanced operations, so those features are planned next. The idea is that you can do:
# create reference objects to on-disk fst files
ft1 <- fst("1.fst")
ft2 <- fst("2.fst")
ft3 <- fst("3.fst")
# add rows to a fst files
rbindlist(ft1, list(ft2, ft3))
# add rows to fst file from in-memory tables dt1 and dt2
rbindlist(ft1, list(dt1, dt2))
# create a reference to 3 fst files (virtual row binding)
ft4 <- fst("1.fst", "2.fst", "3.fst")
# do some calculations but still a reference
ft5 <- ft4[, .(A = sum(B)), by = "C"]
# write result to new file
ft5 %>%
write_fst("5.fst")
# the same with dplyr interface
ft4 %>%
group_by(C) %>%
mutate(A = sum(B)) %>%
compute("5.fst")
without the actual implementation it's difficult to make clear but the data.table and dplyr interfaces will point to an on-disk data source (the fst file). Any operation on them will still be a reference with as much information as possible kept in the file.
But interactive printing and selection will be possible to allow working with the data as you would with an in-memory source (only showing small pieces of the table as usual). Only when you use compute() (dplyr interface) or write_fst() (data.table interface), the data is stored in a new file requiring very little memory. That allows the user to work with data sets much larger than the available memory, while still keeping the interface very responsive.
@MarcusKlik Thanks. Very useful features. I am currently trying to find a solution to "rbind" several large data frames/data.tables into a very large data frame/data.table. I can not find a good memory efficient and fast way to achieve this, besides dumping and appending to a big csv file. Can't wait to try your planned new on-disk features. Any reader or user here have suggestions in the mean time?
Hi @wei-wu-nyc, your specific use-case is interesting because that's exactly the type of problem that I would like fst to help solve (in this case memory shortage).
The most memory efficient way of in-memory row binding is with data.tables rbindlist I guess:
library(pryr)
library(data.table)
# generate sample data.frame
sample_table <- function(id, nr_of_rows) {
data.frame(X = 1:nr_of_rows, Y = runif(nr_of_rows, 0, 100))
}
# generate 20 million row samples and put them in a list
mem_change(
dt_list <- lapply(1:10, sample_table, 20e6)
)
#> 2.4 GB
# row bind them into a single table
mem_change(
dt <- rbindlist(dt_list)
)
#> 2.4 GB
Sys.sleep(5)
# write to disk and clear memory
fwrite(dt, "1.csv")
rm(list = ls())
gc()
# read dt again
mem_change(
dt <- fread("1.csv")
)
#> 2.65 GB
that shows that when you rbindlist a list of data.frame's, you basically need twice the memory of all table's combined. So during the row binding, memory is only allocated for the new vector, which is consistent with the OS memory use:
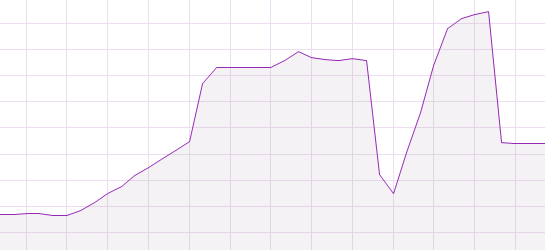
The middle of this graph is the total memory usage after the rbindlist.
You would expect that loading directly from a csv file into memory would take less of your memory, but it doesn't. Although the final memory usage (right side of graph) is equal to the size of dt, during the actual loading of the csv with fread, twice the memory is allocated (right peak). So if the amount of memory is your bottleneck, constructing and then reading from a large csv won't help you, you can just as well rbindlist them in memory.
What would you like to do with the resulting table? Can your data be logically partitioned? Perhaps you only need a few columns for your calculations?
@MarcusKlik What you described is what I observed also. rbindlist() although more efficient than the default rbind(), in the process it still uses close to twice of the final memory footprint. I didn't realize that when reading csv file into memory using fread, the interim memory usage can be twice of the final memory usage. I tested read.fst(), when loading in the previously saved .fst file that contains the large data.table, the memory peak is only slightly bigger than the final memory usage. I try not to split the data into different partitions. We are in the process of cutting down the really needed columns. That may help us to shrink the memory footprint. But your planned feature of on-disk combination of multiple fst files should be very helpful. Microsoft, revolution R has an on-disk data format and a set of functions as well as algorithms that seems to do what I want. But last time I tested, the read and write speed was order of magnitude slower.
Thanks, yes, read_fst only allocates the table once and then just overwrites the elements in-memory, so that should take about the same size as the resulting vector (plus some buffering), so reading directly from a fst file instead of a csv already saves you half of the required memory :-).
Thanks for the pointer to the revolution R algorithms, I will have to study those for sure!
@MarcusKlik I am not too familar with those anymore. But there are set of functions like: rxImport, rxDataStep etc.
I noticed that v0.9.0 was released a month or so ago, and this issue was originally targeted for that release. I see that this particular enhancement is still part of the Candidate milestone. Any further thoughts on when it might be completed?
Hi @phillc73, yes, v0.9.0 was released earlier because of the CRAN issue with rchk.
Apologies for the confusion, this feature should have been moved to the v0.9.2 milestone, thanks for pointing that out!
Hi @wei-wu-nyc, your specific use-case is interesting because that's exactly the type of problem that I would like
fstto help solve (in this case memory shortage).The most memory efficient way of in-memory row binding is with
data.tablesrbindlistI guess:library(pryr) library(data.table) # generate sample data.frame sample_table <- function(id, nr_of_rows) { data.frame(X = 1:nr_of_rows, Y = runif(nr_of_rows, 0, 100)) } # generate 20 million row samples and put them in a list mem_change( dt_list <- lapply(1:10, sample_table, 20e6) ) #> 2.4 GB # row bind them into a single table mem_change( dt <- rbindlist(dt_list) ) #> 2.4 GB Sys.sleep(5) # write to disk and clear memory fwrite(dt, "1.csv") rm(list = ls()) gc() # read dt again mem_change( dt <- fread("1.csv") ) #> 2.65 GBthat shows that when you
rbindlistalistofdata.frame's, you basically need twice the memory of all table's combined. So during the row binding, memory is only allocated for the new vector, which is consistent with the OS memory use:
The middle of this graph is the total memory usage after the
rbindlist. You would expect that loading directly from acsvfile into memory would take less of your memory, but it doesn't. Although the final memory usage (right side of graph) is equal to the size ofdt, during the actual loading of thecsvwithfread, twice the memory is allocated (right peak). So if the amount of memory is your bottleneck, constructing and then reading from a largecsvwon't help you, you can just as wellrbindlistthem in memory.
I think I'm having this issue at the moment. I output several dataframes as part of an analysis (around 1-2 GB each) and tried to bind them all together and save a single output. I hit a memory block at the rbindlist level. I was wondering if something like this should work?:
data <- data.frame(time = 1:2000,
df_name = rep(letters[1], 2000),
v1 = rnorm(2000),
v2 = rnorm(2000))
data1 <- data.frame(time = 1:2000,
df_name = rep(letters[2], 2000),
v1 = rnorm(2000),
v2 = rnorm(2000))
print(object.size(data1), units = "Kb")
print(object.size(as_fst(data1)), units = "Kb")
data_fst <- as_fst(data)
data1_fst <- as_fst(data1)
rbindlist(list(data_fst, data1_fst))
I don't have much understanding of the workings of fst so maybe this is not even possible!
P.S. The fst package has been a MASSIVE help to me for large model outputs. Very much appreciated, I tell everyone about it.
I think the same issue still going on, I checked the newest documentation and didn't see anything about merging on a disk. I also have large data stored on my disk and don't really know how to handle it. Something to solve this issue would be great!
+1
+2
Hello! Tell me please, Is there any plan to support append mode for fst::write_fst()?
I have 0.9.8 version of fst and it seems the append is still not there. I see a lot of people are wating for this killer feature. Could you pleeeease let us know a definite date you plan to implement. I may be using fst only because of this feature.
Status on this ? If you have not gotten around yet, can you point me to where to start implementing this myself. If I understand correctly fst already has everything required to do this, so we could load the pointer to the fst table then call a column append by doing it similar to how your write columns to fst files originally? (I see it does this with a for loop per column). Some questions then arise when I read the code, like would we need to recompress the entire table or only the new columns, since from what I see there is not really any compression between columns anyway ?
I thought a simple test first where I only allow double 64 values to be stored could make it more straightforward since the whole column factory with checks for all input types can be skipped.
Thanks for any help you have on fixing this problem, a lot of people want it I think :)
I was looking for this exact feature, and was hopeful when I saw it was first brought up 5 years ago :-) There is no columnar format that implements this on disk that I know of (poking around arrow suggests you have to create a messy multi-file directory, which is not the best solution IMV). This would be a very powerful enhancement I think, and the main weakness of all popular columnar formats. It's very hard to create output files much bigger than memory, whereas writing out to a bog standard csv allows you to do this trivially, but tonnes of downsides with that solution (which is what we are currently using, alas).