fst
 fst copied to clipboard
fst copied to clipboard
cbind table
Hi I have datasets that is ~1TB for each projects which is too large for a 32GB RAM. I was wondering if there's a way to cbind fst table in a hybrid way?
Basically implement
$<-.fst_table
that append the new column to existing file?
duplicated with #153
Hi @dipterix, thanks for your question!
Yes, the ability to column- and row-bind multiple fst files into a single file is planned but not implemented yet. I'm interested in the details of your specific use-case; is your source data already in a series of fst files, of do you have the data in an alternate source such as a remote database or a batch of csv files?
Most analyses can be split into multiple sub-analyses that each use only a limited amount of columns, which are combined at a later stage. Is there a specific reason why having all data in a single fst file is important for your case?
thanks!
@MarcusKlik I have data in other formats, but also have fst format as cache. For description of my current data, think of time series. My data has 1000 columns, each column is a 1GB signal and can be preprocessed in parallel. However, when I read data, I'm always reading the same rows across the dataset. I thought it might be faster to load these rows if lower end code supports it.
For example, my experiment has 300 trials, each trial ii starts randomly (cannot be determined before slicing, hence I must store the whole data and slice them on the fly), but lasting for 20000 rows. What I want to do is to read 250 columns from all columns each time and reshape to a 300 x 20000 x 250 tensor into RAM.
Right now I store each column as one file and here are two ways I tried to load data:
- For each of these 250 columns, for each trial, load a
20000vector usingfst::read_fst(..., from, to) - For each column, load all 1GB data, slice them into a
300 x 20000matrix
For one single column, the second method is faster. However, when I try to load more columns, R triggers gc(), which takes ~400 ms later on for each columns.
Therefore I switched to the first method, but that's slow too. I guess this is because I have to switch between different files and instead of reading from 250 files, I'm reading from 250 x 300 files?
In summary, my real problem is there's no proper way to load such a large file in the high-end code in R. Either I have to find ways to control gc manually, or I have to seek for low-end implementation/improvement. Also, one-file structure is easier to share and I can always assume those columns exist. Right now if someone removes one file, the reading process (takes minutes) raise errors. It might be their fault removing files, but I think one file is a win-win.
I'll have the speed test results attached later :)
Hi @dipterix, thanks a lot for taking the time to explain your analysis!
The fst format is a columnar format, so reading as many rows as possible during a single read should be the fastest solution (your second option).
From your answer I understand that garbage collection of a single large vector (plus your sliced matrix) takes a relatively long time. Longer than garbage collection of the 250 x 20000 datasets that are loaded with option 1?
Sometimes garbage collection seems to take longer because the RAM is almost fully exhausted. To test that, you could read slightly smaller datasets in memory and force garbage collection after each step (by using gc() after each read). That way, you'll be sure the OS is not using any swap space to increase the amount of virtual memory.
Although having all data in a single fst file will be more convenient as you say (for sharing and completeness), I think the speed gains from accessing a single file as compared to multiple files will be minimal in your particular case.
What would really help you I think is a single buffer that you can use multiple times. Something like:
# write some dataset with 1e8 rows
data <- data.frame(X = sample(1:10, 1e8, replace = TRUE), Y = sample(1:100, 1e8, replace = TRUE))
fst::write_fst(data, "data.fst")
# read first chunk
buffer <- fst::read_fst("data.fst", 1, 1e7)
# do some magic here with the buffer contents
for (chunk in 1:9) {
buffer <- fst::insert_fst(buffer, "data.fst", chunk * 1e7 + 1, (chunk + 1) * 1e7)
# do some magic here with the buffer contents
}
The idea behind fst_insert() would be that existing vectors are overwritten in memory, so no garbage collection will be required between reads, significantly speeding up your experiment (now, the garbage collection takes as much time as the 1 GB reads).
In short, I think when working with such large datasets, row- and column- binding features for writing the data to disk will be very useful. And for reading, a method like fst_insert() could help a lot keeping memory requirements low.
(I realize this won't help you too much now while you need it :-))
Thanks!
That'll be great. On the other hand I was wondering how hard it'll be to use rows instead of having from and to in read_fst and support readings non-consecutive but ordered rows in the C/C++ end?
Like:
fst::read_fst('data.fst', rows = c(1:100, 1004:2000))
Subsetting in C++ might be faster than R and much lighter.
Hi @dipterix, yes, using a row selector during reading will be faster and much more memory efficient. Your question is also related to this issue which is about the use of a row selector for reading to and writing from disk (both are very useful).
That feature request is about random row selectors. For reading, ordered row selectors (like yours) are somewhat easier to implement, because the row index doesn't have to be sorted first before reading from disk.
Going further, it might also be useful to detect ordered sequences (like your 1:100), to be able to read data even more efficiently:
# note: windows only at the moment (some ALTREP functions are not
# exported on linux/OSX), requires R >= 3.6
devtools::install_github("fstpackage/lazyvec", ref = "develop")
# define ordered integer sequence
rows <- 1:100
# for R >= 3.5, this is implemented as an ALTREP vector
lazyvec::is_altrep(rows)
#> [1] TRUE
# with base ALTREP class 'compact_intseq'
lazyvec::altrep_class(rows)
#> [1] "compact_intseq"
Ordered sequences like 1:100 are implemented as ALTREP vectors for R >= 3.5 and can be
detected as such. If fst would read the ALTREP metadata, than e.g.
fst::read_fst('data.fst', rows = 5:100)
can be (internally) converted to
fst::read_fst('data.fst', from = 5, to = 100)
increasing efficiency. Combining multiple ALTREP sequences won't work however:
# combine two ALTREP vectors into a single vector
rows <- c(1:100, 1004:2000)
# the result is not an ALTREP anymore (full expansion)
lazyvec::is_altrep(rows)
#> [1] FALSE
So in your case, a full scan (C++) of the row selector is needed to determine which rows to drop. But as you say, that will still be much faster than the current implementation where a larger vector is read and subsetted afterwards:
fst::fst("data.fst")[c(1:100, 1004:2000), ]
thanks, implementing row-selection during reading and writing is high on the priority list!
For those who are interested in how I compare these two ways of loading data, here are profiling.
Subset data when reading via fst
#### Subset using read_fst(..., from, to)
profvis::profvis({
print(system.time({
vapply(row_start, function(idx){
as.matrix(fst::read_fst(file, from = idx, to = idx + 199))
}, FUN.VALUE = re)
}))
})
#> user system elapsed
#> 0.075 0.027 0.102
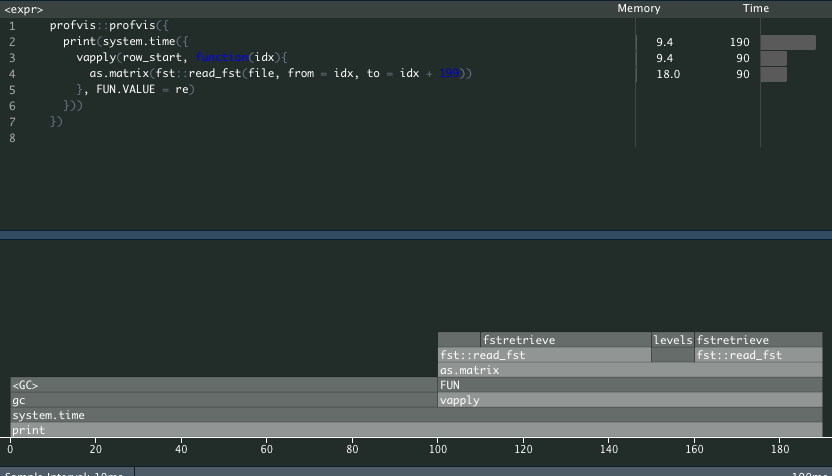
This method almost instantly finished.
Load all data and then subset for each column
In this scenario, suppose I can only hold one column in RAM at a time (for example, I generate a 200GB data on a server with 80 columns and client only has 4GB RAM).
#### Subset in-memory
profvis::profvis({
print(system.time({
lapply(seq_len(10), function(ii){
fst = fst::read_fst(file)
vapply(row_start[ii, ], function(idx){
as.matrix(fst[idx: (idx + 199),])
}, FUN.VALUE = re)
})
}))
})
#> user system elapsed
#> 345.159 63.991 57.580
This one took almost one minutes.
Profiling timer seems incorrect. I guess this is because I ran with 8 threads, so time shows 8x as it should be. gc() is expensive in R (~4 sec total) compared to previous (~20 ms total). And the data I'm targeting is 1000x larger. In addition, memory changes a lot.

Here is how I generated test data.
#### fst reading test
x = rnorm(3e8); dim(x) = c(3e6, 100); x = as.data.frame(x)
pryr::object_size(x)
#> 2.4 GB
#### Create a temp file for testing
file = tempfile(); fst::write_fst(x, file, compress = 100)
#### Generate row indices
row_start = sample(3e6-100, 20)
re = array(0, c(200, 100))
Hi @dipterix, thanks for sharing your benchmarks!
Yes, system.time() seems to take into account the time spent by all the threads, so that's not very useful for multi-threaded code! Package microbenchmark does not have these problems and will yield better results (with more resolution).
From your code, it seems that copying takes a long time. And if memory is almost full, it can take even longer because of R's relatively slow garbage collection. In the test below I compare the speed of reading from a fst file to taking a subset. Copying seems a factor of 2 faster (as you would expect), but I still have plenty of RAM left...
# fst reading test
x = data.frame(X = sample(1:1000, 1e8, replace = TRUE))
file = tempfile()
fst::write_fst(x, file)
random_rows <- sample(1e8, 100)
# this will result in 500 loads
microbenchmark::microbenchmark({
lapply(1:100, function(z) {
y <- fst::read_fst(file, from = random_rows[z], to = random_rows[z] + 999999)
})
}, times = 5)
#> Unit: milliseconds
#> expr
#> { lapply(1:100, function(z) { y <- fst::read_fst(file, from = random_rows[z], to = random_rows[z] + 999999) }) }
#> min lq mean median uq max neval
#> 704.9919 708.8402 838.7643 863.5971 890.9763 1025.416 5
fst_data <- fst::read_fst(file)
# this will result in 500 copies
microbenchmark::microbenchmark({
lapply(1:100, function(z) {
y <- fst_data[random_rows[z]:(random_rows[z] + 999999),]
})
}, times = 5)
#> Unit: milliseconds
#> expr
#> { lapply(1:100, function(z) { y <- fst_data[random_rows[z]:(random_rows[z] + 999999), ] }) }
#> min lq mean median uq max neval
#> 385.3657 434.4636 429.8669 440.5715 441.5131 447.4206 5
It could also be that in your second example, R has more trouble allocating memory because it needs to allocate RAM for the complete dataset. When you read smaller chunks from disk, you only need to allocate smaller RAM sections. And when RAM is almost full, that might take more time.
In the end, I think reading the complete set into memory and then using chunks from that will be slower than just reading the smaller chunks directly. The overhead for file access is small, especially for chunks with medium size.
Perhaps it's possible to write your source data as a single column instead of casting the matrix to a data.frame (a matrix is just a single large vector with some meta-data). That would avoid all the casting and might speed things up?
In my test, I load one column and subset each time. Think if the file is ~200GB with 80 columns (2.5GB per column) and your RAM is only 4GB, you can only load one column at a time. Loading all the data and then subset is the fastest but that's not possible in this scenario, hence fst_data <- fst::read_fst(file) will blow up the RAM.
I think it's the column selection that slow down the whole process.
@MarcusKlik thank you for developing such great package and appreciate for the recently published fstcore package. I solved my issue by implementing cpp level control with fstcore (lazyarray). This issue can be closed.
Hi @dipterix, thanks for the heads up and great to hear that direct usage of the fstcore API works for you!