Error in using Botorch models
Hi, I am using all botorch models(QEHVI, QPAREGO). for my usecase I am registering several metrices in the experiment and 2 as objectives for optimization in optimization config in experiment, However I have found that during the model fit, it uses observation data(all metric values) to fit the surrogate model. Thus my 2 objective optimization problem becomes >>2 optimization problem. Is there a way I can resolve it. I am using ax-platform = 0.2.5.1 version
Thanks for the question @ayushi-3536! I'm not sure this is a problem necessarily. The model fits on all metrics, but will only generate candidates using the optimization config metrics. If you were to plot the predicted outcomes of the generated candidates, you would be able to see the predicted values for the tracking metrics. But it isn't optimizing on the tracking metrics. It doesn't know which direction to optimize in for tracking metrics. Is the problem that you think it's fitting the optimization config metrics differently than it would have if you hadn't passed it tracking metrics as well?
Hi @ayushi-3536, do you still need help with this issue or did @danielcohenlive's comment above fully take care of your issue?
Closing as inactive. Please reopen if following up.
Is the problem that you think it's fitting the optimization config metrics differently than it would have if you hadn't passed it tracking metrics as well?
Hey, im having this issue here using the service API with a fixed seed on the ax client.
When i just register the objective metric ("AdaptedBranin") I get following results:

When i track "other_metric" the results of the objective metric are different. (Notice from the botorch steps) But that shouldn't be the case... or am I wrong there? The model fits on all metrics, but if it only optimizes on the objective, the added tracking metrics should be irrelevant...

Its hard to say exactly what the root cause of this is -- could you provide a minimal repro for us to take a look at? There could be a number of things going on here cc @Balandat
Hmm yeah not obvious what is going on here; Note that this is not the "same" problem from the perspective of the code; under the hood we are fitting either a different kind of model (a batched model where the batch represents the outcome) or two independent models - while that should not affect the modeling itself, it will look different from the perspective of the code path, and so there are probably some other random elements in there (sampling different initializations e.g. for the model fitting parameters) that would cause different results even for a fixed seed (since seed is used for different kinds of random draws under the hood).
A simple repro would be helpful here; though I am not sure I would consider this a bug per se.
Sure, here's the repro
# Define a SingleTaskGP with RBF Kernel
class CustomGP(SingleTaskGP):
def __init__(
self,
train_X: Tensor,
train_Y: Tensor
) -> None:
super().__init__(
train_X=train_X, train_Y=train_Y, likelihood=None,
covar_module=ScaleKernel(base_kernel=RBFKernel()),
mean_module=None,
outcome_transform=None,
input_transform=None
)
# Define synthetic test function and evaluate function for the Service API
test_function = AdaptedBranin()
def evaluate(parameters):
x = np.array([parameters.get(f"x{i+1}") for i in range(test_function.dim)])
tf_botorch = from_botorch(botorch_synthetic_function=test_function)
return {
'AdaptedBranin': (tf_botorch(x), None)
}
# Define generation strategy for the Ax client
generation_strategy = GenerationStrategy(
name='SOBOL+CustomGP+Defaults',
steps=[
GenerationStep(
model=Models.SOBOL
num_trials=5
),
GenerationStep(
model=Models.BOTORCH_MODULAR,
model_kwargs={"surrogate": Surrogate(CustomGP)},
num_trials=-1
)
]
)
#Setup ax client
ax_client = AxClient(random_seed=42, generation_strategy=generation_strategy)
ax_client.create_experiment(
name="adapted_branin",
parameters=parameters,
objectives={'AdaptedBranin': ObjectiveProperties(minimize=True)},
tracking_metric_names=['other_metric']
)
# Run the trials evaluating function "adaptedBranin" and geting the prediction of the model to register it as 'other_metric'
for i in range(20):
parameters, trial_index = ax_client.get_next_trial()
model = ax_client.generation_strategy.model
obs_feat = ObservationFeatures(parameters=parameters)
if isinstance(model, TorchModelBridge):
prediction = predict_at_point(model, obs_feat, ('AdaptedBranin'))
prediction = prediction1[0].get('AdaptedBranin')
else:
prediction = 0.0
ax_client.complete_trial(trial_index=trial_index, raw_data={**evaluation_function(parameters), 'other_metric':(prediction, None)})
when I track the metric like this, I get this results for the objective Metric AdaptedBranin

when i dont track the metric (erasing it from the ax_client and raw_parameters) i get the following results
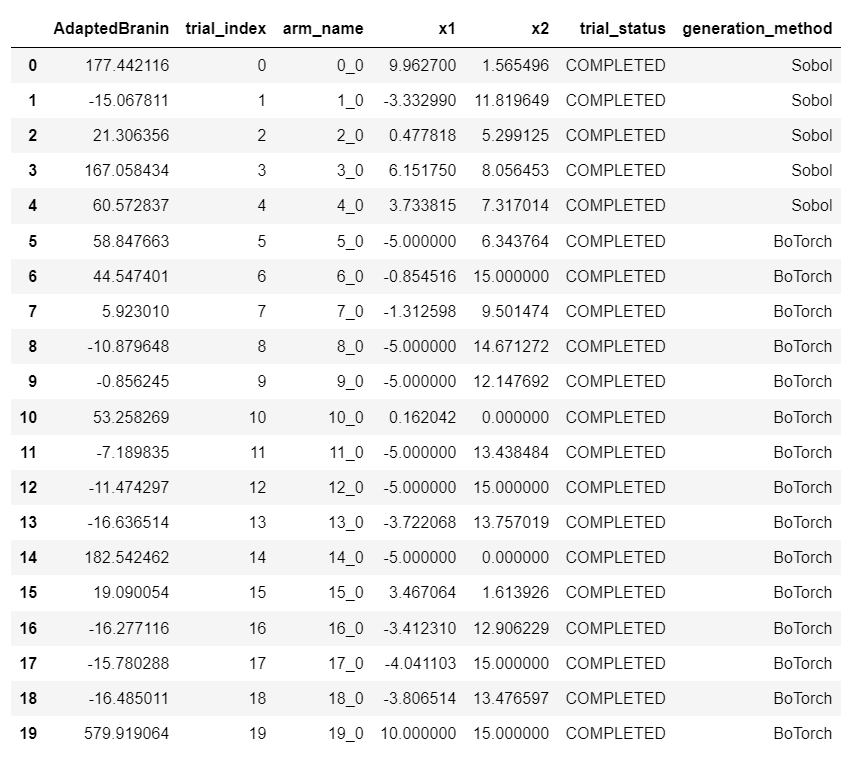
I've tried it with different scenarios comparing the objective metric when tracking the other metric or not:
- without setting the generation strategy (that means taking the default SOBOL+GPEI) it doesnt change.
- using the Models.BOTORCH_MODULAR without any kwargs it also doesn't change.
- using the Models.BOTORCH_MODULAR with surrogate=Surrogate(SingleTaskGP) it also doesn't change. ... I'm I missing somwthing while defining the CustomGP class?
@Balandat, @mpolson64 thank you very much in advance!!
@saitcakmak You have looked into the determinism of candidate generation in the past - do you happen to know what might cause this? No need to go deep if nothing comes to mind, I can take a look as well.
My guess is that this happens because we fit a model to all metrics then subset the ones on the objective to pass to the acquisition function. Since the model fitting spawns a different number of models and consumes a different amount from the RNG stream, the acquisition function ends up being evaluated with different random initial conditions, leading to different candidates being generated and evaluated.
We had discussed implementing a fit_tracking option to allow skipping model fitting for tracking metrics. Something like that would probably resolve this.
Regarding more general determinism: We still have some non-determinism between different machines due to the way pandas sorts the observations, leading to different ordering of train inputs passed to the BoTorch model (which then leads to candidates diverging differently due to various numerical issues). @j-wilson has a change in one of his diffs that deterministically sorts the candidates before passing to BoTorch, fixing this issue. It'd be good to split that up from the larger diff and fix the determinism issue.
Closing this since #1416 is tracking the proposed solution.