tf-faster-rcnn
 tf-faster-rcnn copied to clipboard
tf-faster-rcnn copied to clipboard
A simple idea to solve the error : "rpn_loss_box: nan"
There is an error with bounding boxes' coordinates,when some of coordinate values become 65535.
So, we can open the file named "minibatch.py" , find the line:
gt_boxes = np.empty((len(gt_inds), 5), dtype=np.float32)
and insert some codes in front of it :
boxes = roidb[0]['boxes'][gt_inds, :]
for idx in range(len(boxes)):
boxes[idx][boxes[idx]==65535] = 0
这可能算不上是个好的解决方案 ~
the underlying reason might be your dataset is 0-based rather than 1-based in coordinates?
I faced that issue when forget about -1 for x_max and y_max in dataset loader

I also get this issue. And I have deleted the -1. But it still happen! Does it have a better solution now?

Before running clean your annotation files by checking these conditions. if int(ymin) > int(height): if int(ymax) > int(height): if int(xmin) >= int(xmax): if int(ymin) >= int(ymax): if int(xmin) > int(width): if int(xmax) > int(width) : if int(xmin) <=0 if int(ymin) <=0 if int(xmax) <=0: if int(ymax) <=0:
@FMsunyh Did you delete the .pkl files created in in the cache folder of the dataset? For me deleting the -1 didn't have any effect too at first. But deleting the -1 and after that the .pkl files solved it for me.
@kinzzoku yes, I've deleted .pkl file and rebuild it again, but same issue with me.
There is an error with bounding boxes' coordinates,when some of coordinate values become 65535.
So, we can open the file named "minibatch.py" , find the line:
gt_boxes = np.empty((len(gt_inds), 5), dtype=np.float32)and insert some codes in front of it :
boxes = roidb[0]['boxes'][gt_inds, :] for idx in range(len(boxes)): boxes[idx][boxes[idx]==65535] = 0这可能算不上是个好的解决方案 ~ boxes[idx][boxes[idx]==65535] = 0 请问这样做是为什么?
I have solved this problem .Thank you!
Hello.I have the same problem.
I changed the code as described in the following figure.
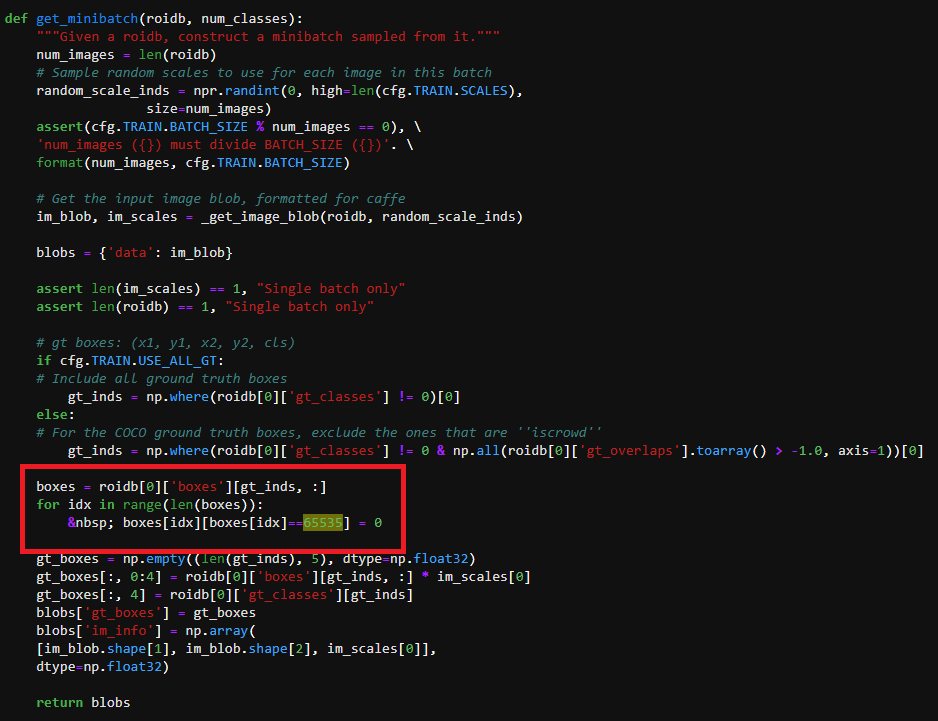
The first error was caused by  .
The error picture is as follows:
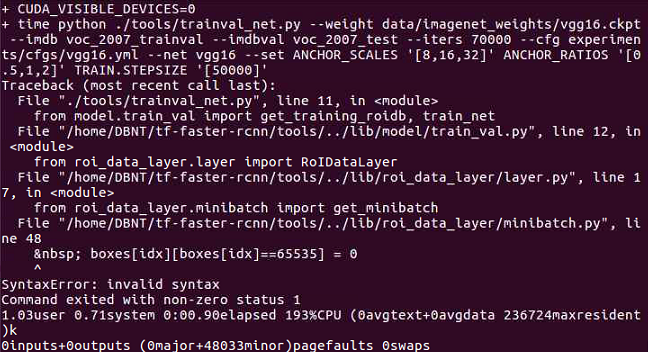
So I erased $nbsp and compiled the code.
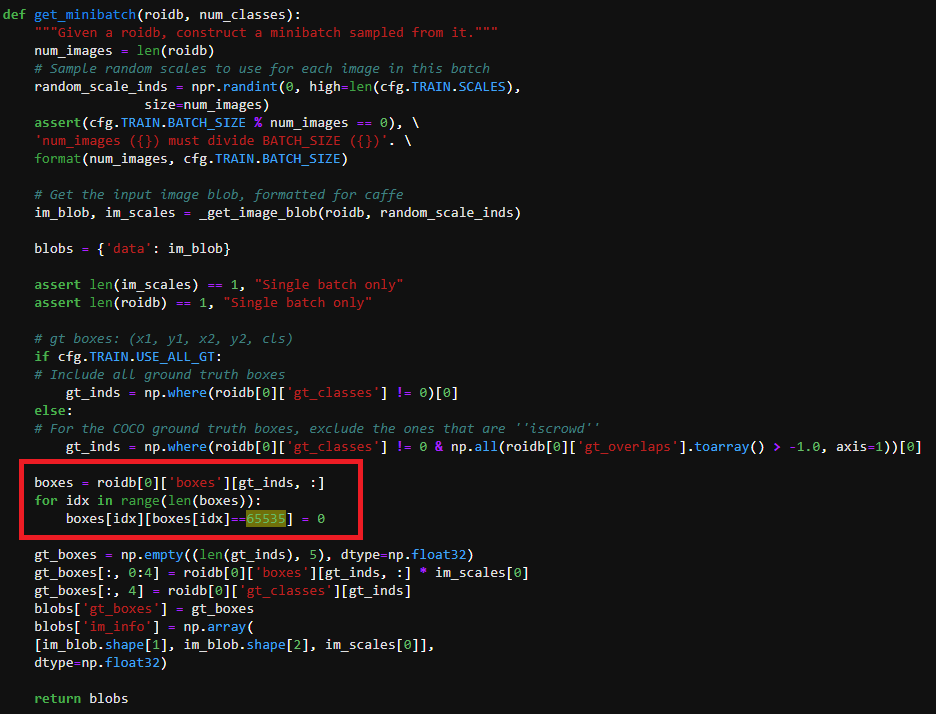
However, a nan value is still generated.
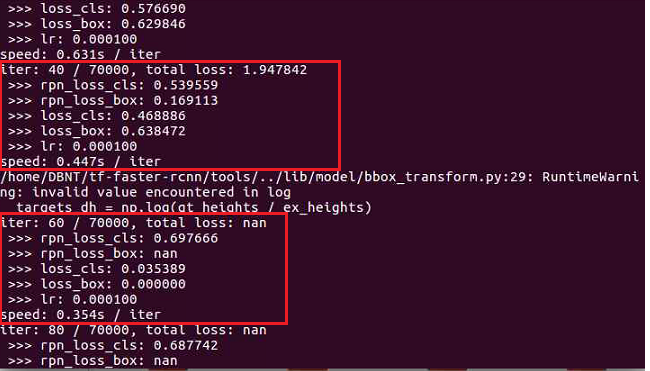
Please help me with the solution.
@NaJongHo hi, I got the same problem. Did you fix it? could you please help me?