Error in solveIRWLS.weights
Hi,
I was running RCTD on multimode and max_cores was set to 1 in order to prevent parallel error. I have 4 ST datasets and in same format, 3 was succesful while one encountered with this error:
Error in S[, i] : subscript out of bounds
Calls: run.RCTD ... process_bead_multi -> decompose_sparse -> solveIRWLS.weights
Execution halted
Could you give me a hint on how to solve this? Thanks a lot
I'm not sure but if you send me the object and script, then I can try reproducing the error.
Dylan
@dmcable Hi Dylan,
The object was uploaded to GoogleDrive here: https://drive.google.com/file/d/16SYzJbtYkWm7CVeZLUe8xNQUg9jcIIyq/view?usp=sharing
and the codes are
library(spacexr)
load('myRCTD.Rdata')
myRCTD <- run.RCTD(myRCTD, doublet_mode = 'multi')
Thanks for your help! Zeyu
Hi, I have tracked the issue down to a problem with small numbers of total counts on these pixels (see attached image below). I have updated the code to throw an error in such a scenario, and to filter out such pixels in the creation of the object. It is worth thinking in your case why these pixels appeared.
You can see that for pixel 414, the total counts is 2, despite the UMI being 199:
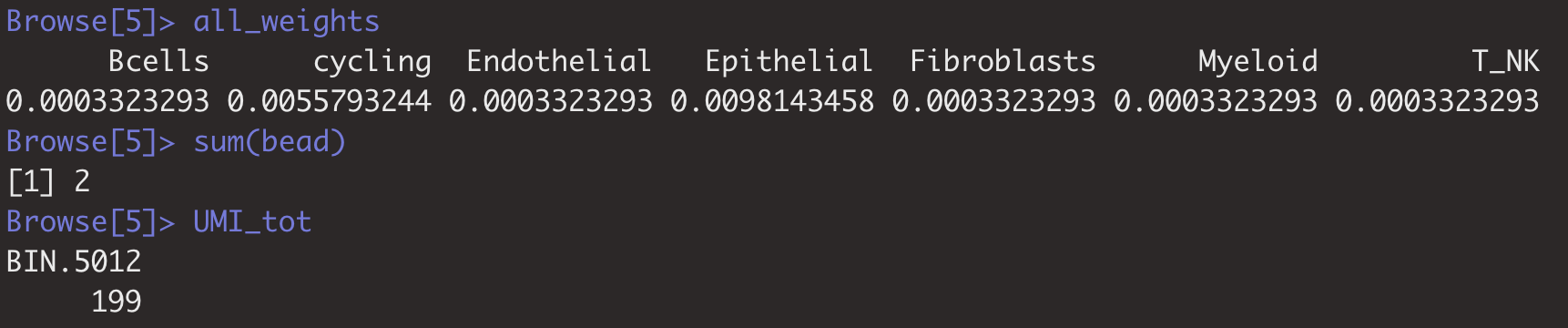
I would recommend filtering out these pixels, which is now default behavior.
I tried again on the latest version but met the error below,
Begin: process_cell_type_info
process_cell_type_info: number of cells in reference: 47950
process_cell_type_info: number of genes in reference: 30109
Bcells cycling Endothelial Epithelial Fibroblasts Myeloid
10000 895 1792 5263 10000 10000
T_NK
10000
End: process_cell_type_info
create.RCTD: getting regression differentially expressed genes:
get_de_genes: Bcells found DE genes: 108
get_de_genes: cycling found DE genes: 161
get_de_genes: Endothelial found DE genes: 203
get_de_genes: Epithelial found DE genes: 275
get_de_genes: Fibroblasts found DE genes: 250
get_de_genes: Myeloid found DE genes: 221
get_de_genes: T_NK found DE genes: 106
get_de_genes: total DE genes: 1169
create.RCTD: getting platform effect normalization differentially expressed genes:
get_de_genes: Bcells found DE genes: 218
get_de_genes: cycling found DE genes: 445
get_de_genes: Endothelial found DE genes: 428
get_de_genes: Epithelial found DE genes: 547
get_de_genes: Fibroblasts found DE genes: 415
get_de_genes: Myeloid found DE genes: 398
get_de_genes: T_NK found DE genes: 300
get_de_genes: total DE genes: 2222
fitBulk: decomposing bulk
chooseSigma: using initial Q_mat with sigma = 1
Likelihood value: 209006.440472752
Sigma value: 1.16
Likelihood value: 207218.854991254
Sigma value: 1.32
Likelihood value: 206087.778346392
Sigma value: 1.48
Likelihood value: 205524.395898714
Sigma value: 1.58
Likelihood value: 205421.245595721
Sigma value: 1.58
Error in process_bead_multi(cell_type_info, gene_list, puck@nUMI[i], beads[i, :
process_bead_multi: no cell types passed weight threshold on full mode. Please check that enough counts are present for each pixel
Calls: run.RCTD ... fitPixels -> process_beads_multi -> process_bead_multi
Execution halted
I wonder what does the "bead" mean here? Is that meaning the number of celltypes or what? Besides, what does the "initial_weight_thresh" stand for? Could you explain it for a bit? Thanks again! @dmcable
Yes, so now this error indicates that something is wrong. Bead is synonymous with pixel (or spot). The initial_weight_thresh, set to .01 by default, is the amount of weight a cell type must have in full mode in order to be considered for multi mode. In this case, the weight of all cell types did not pass threshold. You can proceed by removing the cell types with small total counts, which is done automatically by the updated create.RCTD function.