What does the parameter "thin" mean?
General information:
- emcee version: 3.0.2
- platform: Ubuntu
- installation method (pip/conda/source/other?): pip
Problem description:
I'm wondering what does the parameter 'thin' exactly mean? I find for different thin value, the result is different.
So I'm confused what value should I set to the parameter 'thin': flat_samples = sampler.get_chain(discard=100, thin=?, flat=True) ?
let steps= 5000, walks=32, discard=100, and let
flat_samples = sampler.get_chain(discard=100, flat=True, thin=i)
If I denote the size of flat_samples when thin= i as ' flat_samples| thin= i ', I found:
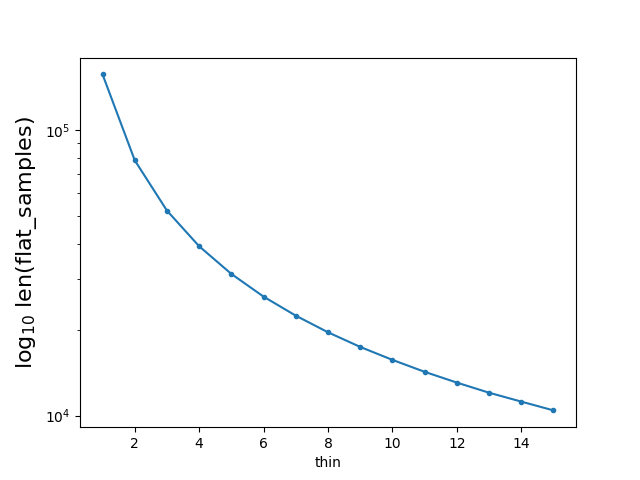
flat_samples | thin=1 = (5000-100)*32 =156800
flat_samples | thin=i = [ (5000-100)/thin] * 32
But I still do not understand the meaning of the parameter 'thin'. Why should one 'thin' a relatively big sample to a small sample?
Code to show my question:
import numpy as np
import matplotlib.pyplot as plt
np.random.seed(123)
# Choose the "true" parameters.
m_true = -0.9594
b_true = 4.294
f_true = 0.534
# Generate some synthetic data from the model.
N = 50
x = np.sort(10 * np.random.rand(N))
yerr = 0.1 + 0.5 * np.random.rand(N)
y = m_true * x + b_true
y += np.abs(f_true * y) * np.random.randn(N)
y += yerr * np.random.randn(N)
def log_likelihood(theta, x, y, yerr):
m, b, log_f = theta
model = m * x + b
sigma2 = yerr ** 2 + model ** 2 * np.exp(2 * log_f)
return -0.5 * np.sum((y - model) ** 2 / sigma2 + np.log(sigma2))
def log_prior(theta):
m, b, log_f = theta
if -5.0 < m < 0.5 and 0.0 < b < 10.0 and -10.0 < log_f < 1.0:
return 0.0
return -np.inf
def log_probability(theta, x, y, yerr):
lp = log_prior(theta)
if not np.isfinite(lp):
return -np.inf
return lp + log_likelihood(theta, x, y, yerr)
x0=np.array([-1.00300851, 4.52831429, -0.79044033])
import emcee
pos = x0 + 1e-4 * np.random.randn(32, 3)
nwalkers, ndim = pos.shape
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_probability, args=(x, y, yerr))
sampler.run_mcmc(pos, 5000, progress=True);
flat_samples = sampler.get_chain(discard=100, thin=15, flat=True)
print(flat_samples.shape)
x=np.zeros(15)
y=np.zeros(15)
for i in range(1,16):
flat_samples = sampler.get_chain(discard=100, flat=True, thin=i)
x[i-1]=i
y[i-1]=len(flat_samples)
fig, ax = plt.subplots()
ax.semilogy(x, y,'.-')
plt.xlabel("thin")
plt.ylabel(r"$\log_{10}$ len(flat_samples)",fontsize=16);
plt.savefig("thin.png")
plt.show()
# sample code goes here...
By running the above code, I got a figure showing the relation between the value of thin and len(flat_samples):
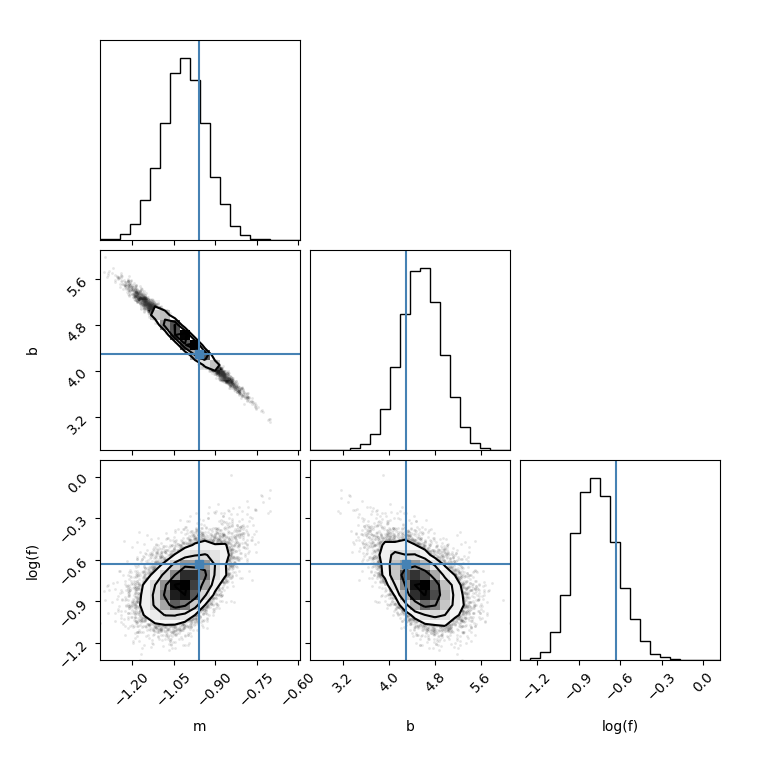
Then I made a corner plot by setting thin=1
flat_samples = sampler.get_chain(discard=100, thin=1, flat=True)
import corner
labels = ["m", "b", "log(f)"]
fig = corner.corner(
flat_samples, labels=labels, truths=[m_true, b_true, np.log(f_true)]
);

I made another corner plot by setting thin=15, I found the figure is different:
flat_samples = sampler.get_chain(discard=100, thin=15, flat=True)
Then I run the following code, I found for different thin value, the results is also different.
from IPython.display import display, Math
for i in range(ndim):
mcmc = np.percentile(flat_samples[:, i], [16, 50, 84])
q = np.diff(mcmc)
txt = "\mathrm{{{3}}} = {0:.3f}_{{-{1:.3f}}}^{{{2:.3f}}}"
txt = txt.format(mcmc[1], q[0], q[1], labels[i])
display(Math(txt))
The samples in an MCMC chain are not independent (see https://emcee.readthedocs.io/en/stable/tutorials/autocorr/ for example) so it can be redundant to store many non-independent samples. thin=n takes the chain and returns every nth sample and you could set n to something like the integrated autocorrelation time without loss of statistical power. You will find that you get marginally different results because MCMC is a stochastic process and you'll see the Monte Carlo error.
tau = sampler.get_autocorr_time()
thin = int(0.5 * np.min(tau))
In the future, I will use the above methods to find the 'best' thin value. Am I right?
No, that's not necessarily right and there's never going to be any "best" thin value. That can be a fine choice in some cases, but remember that the integrated autocorrelation time depends on the target integral so you might be throwing away information for some targets if you use that. I normally don't use the thin parameter, unless I'm storing a large number of chains and want to save on hard drive space.