deepTools

deepTools copied to clipboard
deeptools bam and bigwig peak not the same
Welcome to deepTools GitHub repository! Before opening the issue please check that the following requirements are met :
- [x] I searched and found I encountered exact the same issue in #92, but parameter
fragmentLengthis not used any more.
bamCoverage: error: unrecognized arguments: --fragmentLength 0
- [x] Paste your deepTools version (
deeptools --version) and your python version (python --version) below.
deeptools 3.5.1
Python 3.7.0
- [x] Paste the full deepTools command that produces the issue below (ignore if you simply spotted the issue in the code/documentation).
$ bamCoverage --bam BMMC_donor1_cluster6.bam \
--outFileName bw/BMMC_donor1_cluster6.bin1.bigWig \
--outFileFormat bigwig \
--binSize 1 \
-p 12
- [x] Paste the output printed on screen from the command that produces the issue below (ignore if you simply spotted the issue in the code/documentation).
bamFilesList: ['BMMC_donor1_cluster6.bam']
binLength: 1
numberOfSamples: None
blackListFileName: None
skipZeroOverZero: False
bed_and_bin: False
genomeChunkSize: None
defaultFragmentLength: read length
numberOfProcessors: 12
verbose: False
region: None
bedFile: None
minMappingQuality: None
ignoreDuplicates: False
chrsToSkip: []
stepSize: 1
center_read: False
samFlag_include: None
samFlag_exclude: None
minFragmentLength: 0
maxFragmentLength: 0
zerosToNans: False
smoothLength: None
save_data: False
out_file_for_raw_data: None
maxPairedFragmentLength: 1000
When I load the raw bam file and the output BigWig file into IGV, I find some peaks are different. Like this one:
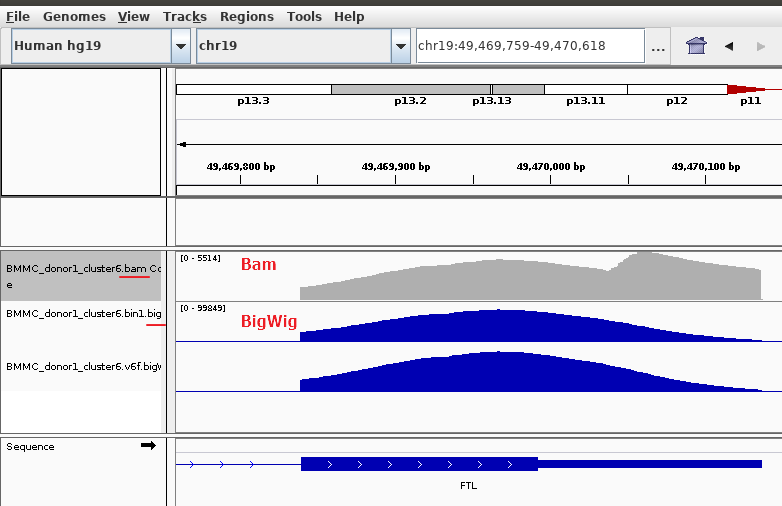
This is a small bam file I sampled from that big bam, which can make the same peak as shown in the picture above.
cluster1.region.0.05.zip
OS: Ubuntu 20.04 reference: hg 19 region: chr19:49469332-49470390
Bye the way, the tdf files( a kind of wig file?) produced by igvtools count, show the same peaks as bam files.
Files:
-rw-rw-r-- 1 wangjl wangjl 14K May 27 11:14 cluster1.region.0.05.count.tdf (-z 1)
-rw-rw-r-- 1 wangjl wangjl 13K May 26 22:40 cluster1.region.0.05.bw (--binSize 1)
-rw-rw-r-- 1 wangjl wangjl 25K May 26 22:29 cluster1.region.0.05.bam.bai
-rw-rw-r-- 1 wangjl wangjl 85K May 26 22:29 cluster1.region.0.05.bam
Version:
$ bamCoverage --version
bamCoverage 3.5.1
$ /usr/bin/java -version
openjdk version "11.0.15" 2022-04-19
## igvtools: 2.5.3
Cmd:
$ bamCoverage --bam bam/test/cluster1.region.0.05.bam \
--outFileName bam/test/cluster1.region.0.05.bw \
--outFileFormat bigwig \
--binSize 1 \
-p 5
$ /usr/bin/java -Djava.awt.headless=true -Xmx1500m \
--module-path=/home/wangjl/anaconda3/share/igvtools-2.5.3-0/lib @/home/wangjl/anaconda3/share/igvtools-2.5.3-0/igv.args \
--module=org.igv/org.broad.igv.tools.IgvTools count \
-z 1 -w 1 bam/test/cluster1.region.0.05.bam bam/test/cluster1.region.0.05.count.tdf hg19