Sinusoidal noise / clusters with multiple peaks
Hello KiloSort team,
I've tried to cluster my data from a freely moving rat with kilosort and found it's fantastic. I really thank you for developing and sharing such great software. Kilosort does great job but I run into the following problems. I would appreciate any helps, suggestions, and comments on these issues.
-
Most clusters have huge sinusoidal noises. Here is an example.
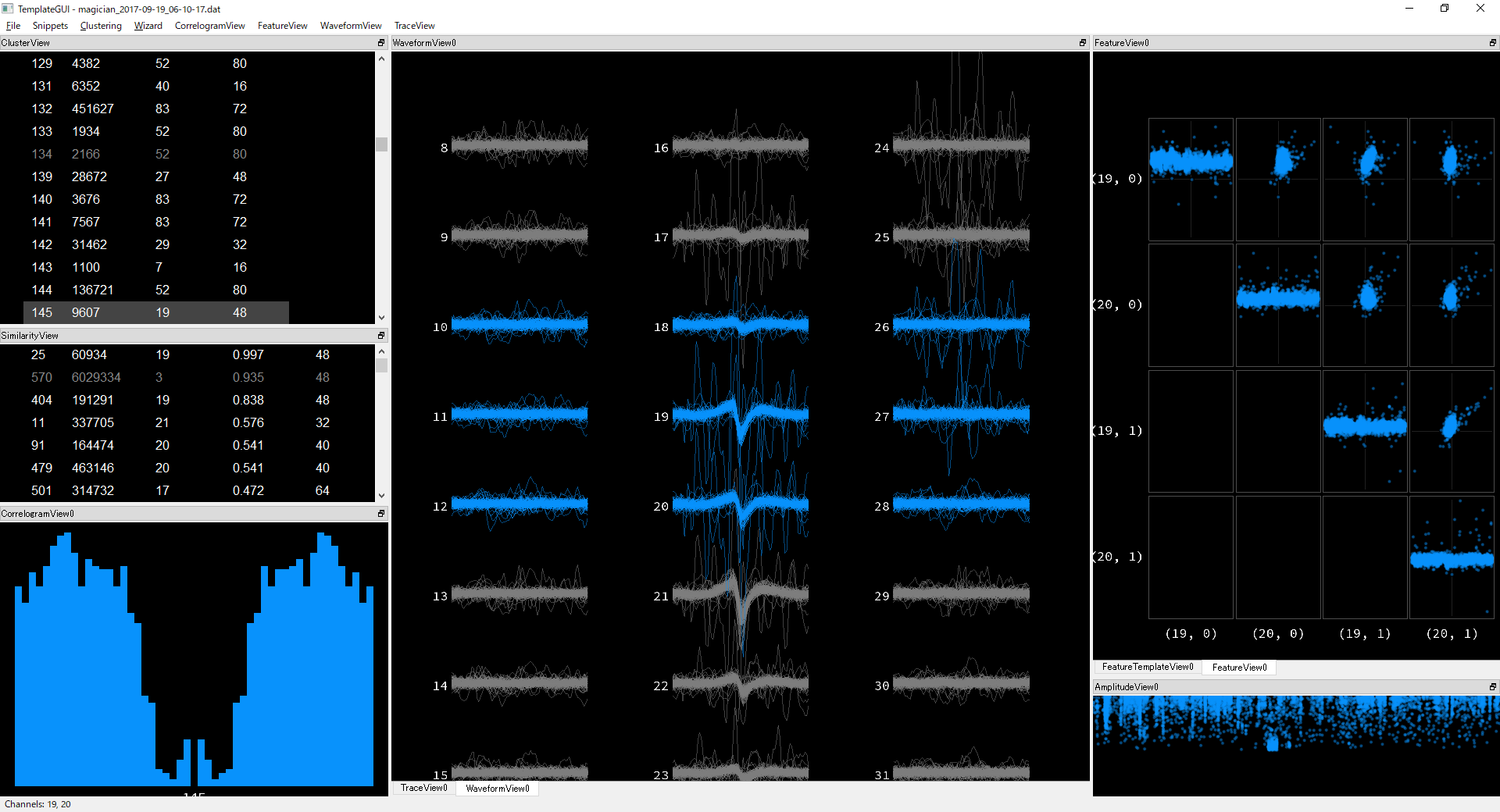 After manual separation, I found actually there are more noise than expected from the plots.
After manual separation, I found actually there are more noise than expected from the plots.
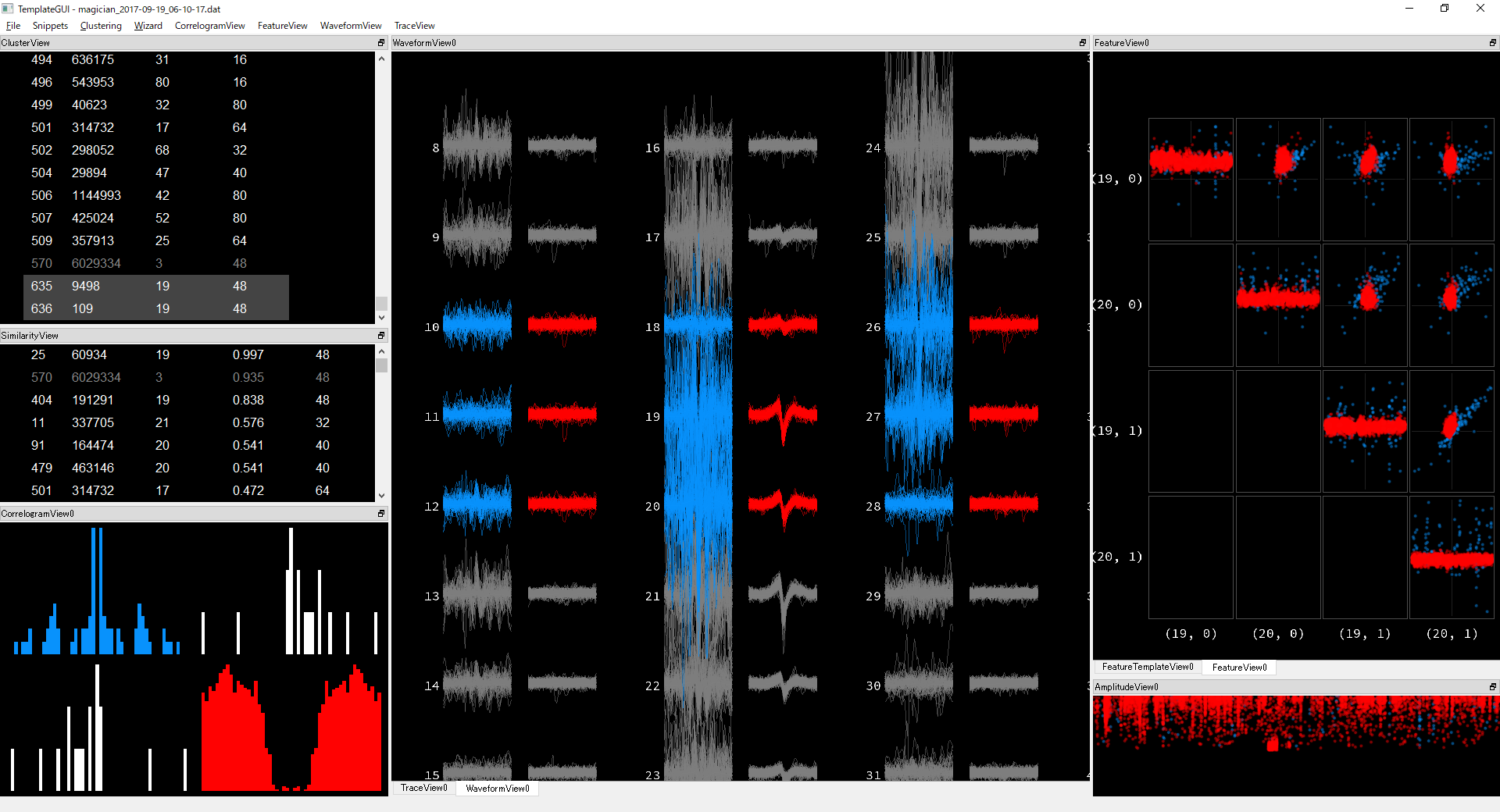 I'm trying to remove them automatically based on amplitudes or PCA scores, but I haven't found a method with satisfying results. Do you know a good way to remove and/or prevent having such kind of noise?
I'm trying to remove them automatically based on amplitudes or PCA scores, but I haven't found a method with satisfying results. Do you know a good way to remove and/or prevent having such kind of noise? -
Spike doublets Sometimes ACG of spike timing has sharp peak at time zero (sorry I don't have examples with me now). I looked closely what causes this, and realized some spikes counted twice. In some cases one template pick one spike twice, and in other cases multiple templates picks the same spike. Is there a good way to deal with this issue?
-
Multiple peak clusters Non-ignorable number of clusters have double peaks.
 Ideally this should be deconvolved to two spikes. And at least this case, another template should have taken the first spike.
Ideally this should be deconvolved to two spikes. And at least this case, another template should have taken the first spike.
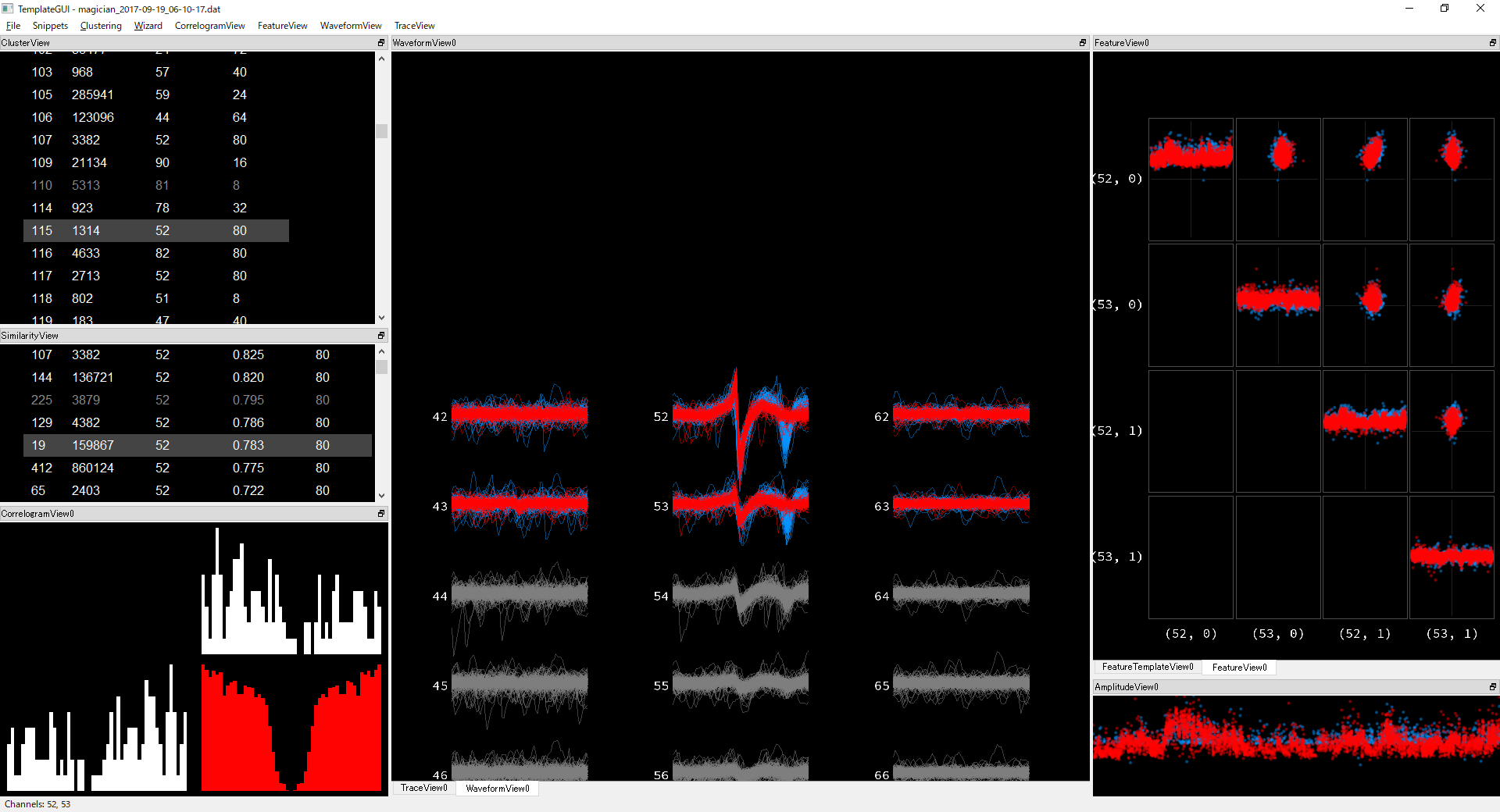 Can I solve this issue by setting parameters properly? Or is there good post-hoc method to fix this?
Can I solve this issue by setting parameters properly? Or is there good post-hoc method to fix this? -
Wrong coloring of waves on phy It's a kind of minor issue, but colored channels are not corresponding to channels of spikes, as you can see on images above. Automatic zoom works properly, so it doesn't cause problem, but if coloring works fine it would be better. Is that an issue on phy, or am I having wrong parameter files?
Thank you.
Hi Hiro,
-
It looks like those are artifacts, such as you get with moving animals, chewing, etc. The problem with these is they are not so huge to be discarded: they are kind of on the order of the template sizes, so they are assigned to templates. They are rare, which is why you only see a few examples when you select a cell. You see more examples when you split it, because Phy always shows a random, small sample of spikes, not all spikes. One thing that could help with this is increasing ops.lam. That is a parameter that ensures spikes have close the same amplitude within cluster. A better fix would require more involved development on my part. I will open another issue to remind myself of it.
-
The ACG doublets are, again, a rare occurrence, relative to the total number of spikes, but it's very obvious in the autocorrelogram plots, which is why you picked it up. You could fix somewhat by increasing the threshold ops.mu for picking up a waveform. However, the number of doublets will have a minimal impact on any further analysis, and you can just post-process to remove them anyway.
-
That you can fix by reducing the number of samples in your waveforms, say to 41 rather than 61. I think that parameter is ops.nt0.
-
This could have to do with the channel map, so check that and make sure it follows the instructions for creating it in createChannelMap.
Hi Marius,
Thank you for comments and advice, and opening new issue for artifacts.
Regarding channel map, I think I have correct x- y- and k-coords, and templates in rez.mat seem to take k-coords into account correctly. Also chanMap, connected, and chanMap0ind are set properly. But somehow colored waves appear strange. An example of a double-peaked cluster shown above has colored waves on ch42, ch43, and so on, but I don't see spikes on them, and on ch54 has gray trace only although it has spikes. Do I need to give additional parameter to channel map?
Hi,
we share Hiro's issues 2 and 3.
-
We do have a lot of doublets in our data. Most clusters in our hippocampal recordings share spikes, like you can see below, And this could be an issue for our sequence analysis. I think that we will write a script to get rid of them and assign every given spike to its most similar cluster
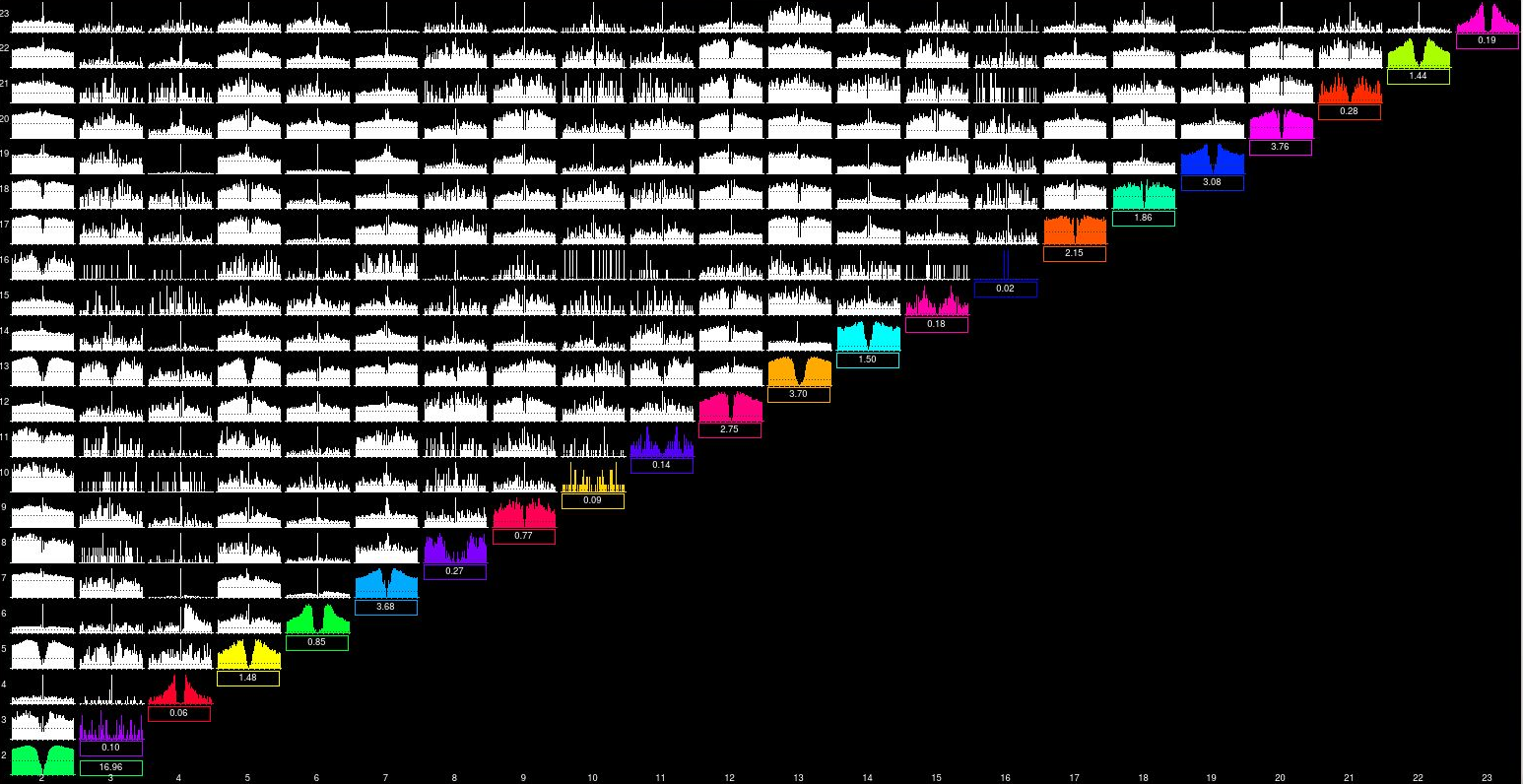 I could not find the parameter ops.mu anywhere. Marius, are you maybe referring to ops.spkTh?
I could not find the parameter ops.mu anywhere. Marius, are you maybe referring to ops.spkTh? -
we have too many multiple peaks clusters so tried many different values of nt0. We have detailed our results in the issue #171 (I'm sorry I did not spot that this issue had already been introduced here). We end up needing an nt0 = 5 to avoid getting any double peak cluster. Does decreasing it so dramatically pose any risks?
Thanks, Marco