PCspikes2 vs PCspikes
Somewhat asked, but not answered, in #78
When visualising the PCs via imagesc of in the Wi variable:
-
PCspikes.mat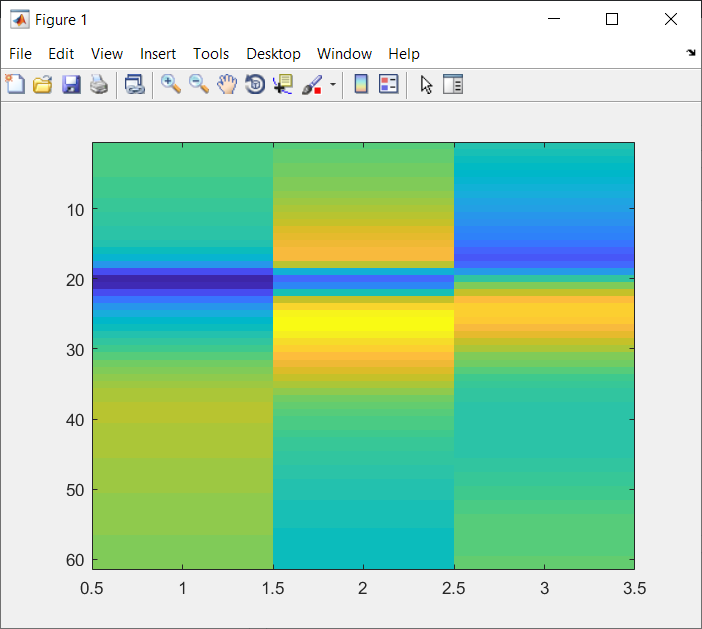
-
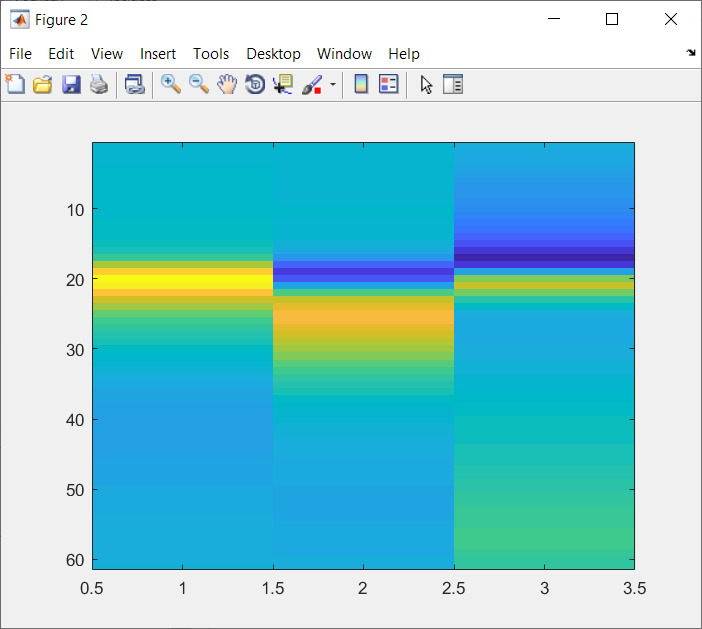
PCspikes2.matOriginal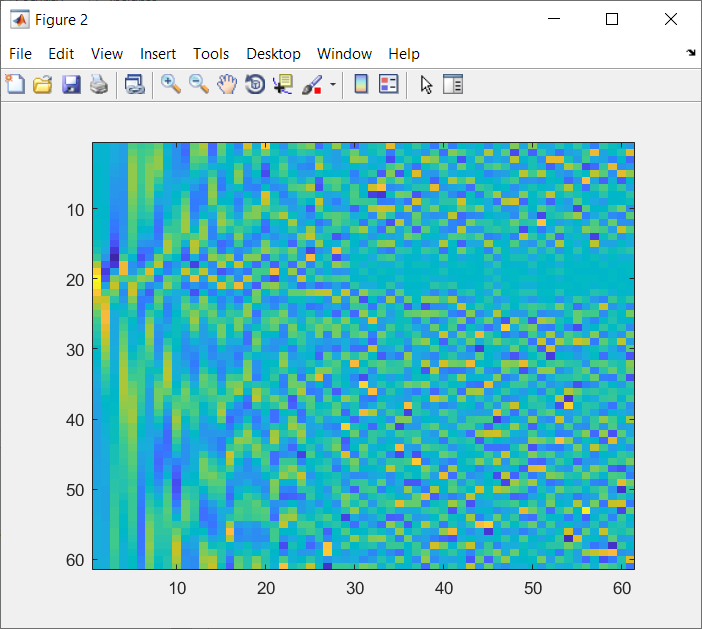 Zoomed into first 3 PCs (to compare with 1. )
Zoomed into first 3 PCs (to compare with 1. )
The question now is: what does PCspikes.mat represent and how can it be computed?
We can see that the PCs in PCspikes.mat are almost inverted compared to those in PCspikes2.mat, but I am unsure if this was a decision or consequence of the computation of these PCs. PCspikes2.mat can be computed as described in: #169.
The Wi can be seen being used in neighbouring PC computation:
https://github.com/cortex-lab/KiloSort/blob/3ff2d1027166b6e54ab4661b8c56e33e1ba9c4e7/fullMPMU.m#L96-L111
and later on during low dimensional projection:
https://github.com/cortex-lab/KiloSort/blob/3ff2d1027166b6e54ab4661b8c56e33e1ba9c4e7/fullMPMU.m#L164
I have also tried the following from another fork:
https://github.com/dimokaramanlis/KiloSortMEA/blob/cdb1bd35d2284f8fb94f7c05d8c18c8250677a3e/fullMPMU.m#L98
Wi=ops.wPCA; %load PCspikes;
The effect of this change is seen in FeatureView and AmplitudeView, default on the left, the above change on the right:
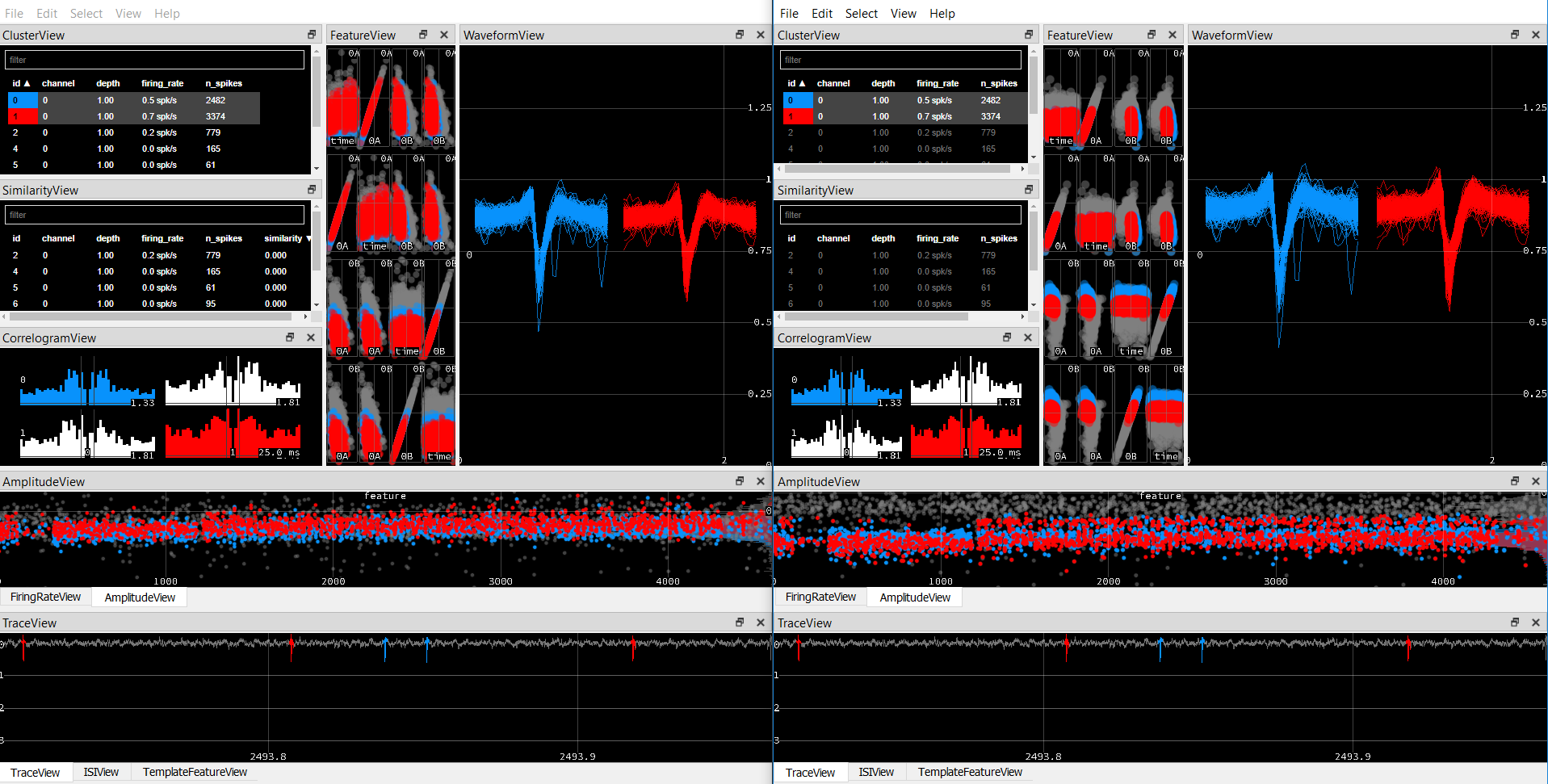
But in our case, we are fairly confident that both of these clusters are contaminated and there are at least 2 distinct waveforms. So although the Feature view seems to make these units more separable in that space, the final cluster allocations are exactly the same.