AutoDock-Vina
 AutoDock-Vina copied to clipboard
AutoDock-Vina copied to clipboard
AutoDock Vina
Hi everyone, I am working on hydrated docking with large ligand library using same script. Hydrated docking works fine for some ligands that do not contain OA or HD atom...
Hi, I use 'prepare_ligand4.py' to convert small molecules from mol2 to pdbqt. I found some molecules changed their structures after converting to pdbqt, for example: 1.mol2: 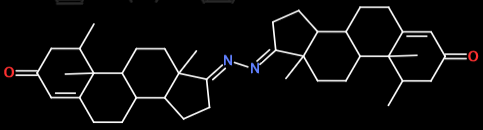 after convert (1.pdbqt):...
Hi, I've tried the following commands to install vina, taken directly from the [Installation Tutorial](https://autodock-vina.readthedocs.io/en/latest/installation.html#installation). ``` $ conda create -n vina python=3 $ conda activate vina $ conda config --env...
Hello, When I was trying to preparing my ligand by adding water molecules, it added too many waters and some of them are too close. When I open the pdbqt...
Dear all, When I generate poses as so: ``` vina --ligand ligand.pdbqt --receptor receptor_rigid.pdbqt --flex receptor_flex.pdbqt --out global_search.pdbqt --cpu 8 --size_x 22.5 --size_y 22.5 --size_z 22.5 --center_x 36.288 --center_y -8.757...
Will be quite helpful and probably, no downsides?
**Issue and aim** I have an issue with the Vina tutorial for rigid receptors: my outputs for the prepare_receptor, mk_prepare_ligand.py and prepare_gpf.py give different results compared to the same files...
Hello Sir Can I use Vina to dock analogues of nucleotides triphosphates into a polymerase active site coordinating Mg ions? The ions are also typically in coordination with water molecules...
Hi, I used two types of AutoDock-Vina; 1. release v.1.2.3 in (https://github.com/ccsb-scripps/AutoDock-Vina/releases) 2. download source code in (https://github.com/ccsb-scripps/AutoDock-Vina.git, currently being updated??) and set-up >> It was also version v1.2.3. The...
I got this error massage when I used the python vina==1.2.3: ``` ERROR: The ligand is outside the grid box. Increase the size of the grid box or center it...
