montepython_public
 montepython_public copied to clipboard
montepython_public copied to clipboard
Positive-Definite Parameters
I've been doing some comparison runs between MontePython and Cosmomc using the Planck 2018 data release, and have been having problems getting constraints to match between the two of them, particularly on the tensor/scalar ratio. I am running with the same likelihoods, the same parameter sets, and the same priors for both.
Looking into it a bit, I noticed that Cosmomc is (correctly) limiting the parameter $r$ to be positive-definite, but MontePython appears to be sampling the region $r<0$. See attached triangle plots, both produced with GetDist. The input distparam files are identical, the only difference is the chains. Note that the cosmomc contours have a sharp cutoff at $r=0$, but the MontePython contours do not. In particular, for t$r$, here is my line from the parameter file, specifying a minimum value of $r=0$:
data.parameters['r'] = [0.06, 0.0,0.5, 0.04,1, 'cosmo']
This appears to be being ignored. Any idea how to remove this undesirable behavior in MontePython? It makes a meaningful difference in the constraints.
Dear Will, Of course, since you have correctly given a min of r>0 in our input file, the region r<0 is not explored by the sampler and all points in the chain have r>0. So this sounds like a plotting issue. The MontePython plotting routine (montepython.py --info) actually parses the input file of the run, or more precisely, it parses the log.param that has been created by the run in the chain directory. The routine reads the min and max value of parameter, and it uses this information to infer that there is 'hard boundaries' and a 'single-sided tail'. The information is then used when defining the bin edges, and it leads to a sharp boundary, like you would expect. Finally the upper boundary will coincide with the one from CosmoMC (as we tested many many times over many years). Thus I guess that the plot you sent was not produced with montepython.py --info? What did you use to produce it? If it were montepython.py --info, I would be very surprised: we have done run with r>0 millions of time and never got such a problem. I would then suggest that you send us a .tar.gz of the whole chain directory, to let us reproduce the problem. Maybe you removed or modified accidentally your file log.param in the chain directory? If it is GetDist, I am not sure, but I think that you have to inform the code manually through the input that the posteriori has a sharp boundary and a single tail. In any case the plotter must get this information independently of the chains themselves, otherwise it would naively create histograms not respecting the r>0 boundary - as in the problematic plot that you sent. Thanks for raising this issue, Julien
Hi Julien --
Thanks! To answer your questions:
-
I produced the plot with GetDist. I'm having a separate problem with montepython --info, which crashes entirely due to a LaTeX formatting error with the BICEP/Keck paramnames, and produces no output. I'm still working on finding a way around that one.
-
My log.param file is present and unmodified.
-
I also expect that this is an issue with postprocessing, but I am correctly specifying a range for $r$ in the distparams.ini file for GetDist. The thing that is confusing me here is that I am postprocessing both the cosmomc chains and the MontePython chains absolutely identically, but only the MontePython-generated chains exhibit the issue, and I'm trying to figure out what the difference is.
Cheers,
Will
I see. In this case the log.param is of course irrelevant, it is not read by GetDist. Then, what I suspect is that GetDist is conditioned to read some kind of meta-data in some file produced by CosmoMC, and for Monte Python chains, it does not find this information, thus it assumes two-tailed distributions. The fact that you told GetDist that r>0 is essential. It seems not to be sufficient. Isn't there an entry in GetDist or in the files produced by CosmoMC where you can specify that the distribution is single-tailed? I was a heavy user of GetDist until 2012, and I remember very well that at that time, there was such a thing... Maybe you don't need to specify it anymore for CosmoMC chains because CosmoMC has already stored this information in one of these meta-data files (exactly like we do with our log.param) ?
Hi Will,
A dirty fix with the BICEP/Keck parameter names: first save a backup of your log.param and then simply change the names of the offending parameters in the log.param file. As long as you're only analyzing doing this should be fine, MontePython will correctly read the parameters in the order specified in the log.param (ignoring the actual names except for labeling plots and info files). If you want to continue the run to get better convergence, you'd need to replace the log.param with the backup of the original or BICEP/Keck wouldn't recognize the nuisance parameters. This way you should be able to let us know if the contour problem you describe persists when plotting with MontePython, or if the issue is as Julien suspects that GetDist requires meta data.
Best, Thejs
Another note, it may be possible to fix the parameter name error by passing a file --extra plot_files/example.plot where you change the parameter names with the info.to_change line, replacing the offending name with one that can be parsed by analyze.
Thanks guys. I'll give this a shot and get back to you.
Re: using MontePython info, I changed the param names in my .paramnames file
` BBdust BBdust
BBsync BBsync
BBalphadust BBadust
BBbetadust BBbdust
BBalphasync BBasync
BBbetasync BBbsync
BBdustsynccorr BBdustsynccorr
`
But it is still reading the original (bad) LaTeX format from somewhere, and I can't figure out where:
! Undefined control sequence. l.13 ...000000}{20.000000}{\sffamily $BB\alphadust $}
The LaTeX format does not appear on the log.param file.
Running with --extra fails, as follows:
`
./montepython/MontePython.py info ./chains/PlanckBK15_test/ --extra plot_files/example.plot Running Monte Python v3.3.2
Traceback (most recent call last):
File "./montepython/MontePython.py", line 40, in
Note that this is with an unmodified example.plot file.
Hi WIll,
MontePython doesn't actually use the .paramnames file, that's for running with GetDist. For MontePython to read the new parameter names, you'd need to change the log.param file (after making a backup). Or, in the example.plot file you have to change the line info.to_change to make it change the offending parameters, e.g.
info.to_change = {'BBalphadust': 'BBadust'}
where I'm guessing the problem has to do with the automatic conversion of alpha to \alpha, but I'm not sure about that. You'd similarly have to add other parameters that cause problems to the same dictionary.
Best, Thejs
You have to modify the example.plot file before it will work. The line #info.to_change = {'rs_d': 'r^s_d', 'omega_cdm': '$\Omega_\mathrm{m}$'} Should be uncommented and with the parameters changed to the problematic ones.
The LaTeX error is saying that there's some problem with the formatting of the parameter $BB\alphadust $ (I'm going to guess because there's no space between alpha and dust, so LaTeX tries to find a variable \alphadust, which doesn't exist).
MontePython's analyze module will take the parameters from the log.param and try to "texify" them. However, this method isn't foolproof and sometimes fails. The parameter 'BBalphadust' in the log.param file is an example of this.
There are a few different ways to go about fixing this, one is changing the parameter using a plot file (with --extra), which if the info.to_change dictionary is set up correctly will tell the code to change the name of the parameter name before texifying it. I think that should work. Another is to change the name at the source, i.e. in the log.param. Changing 'BBalphadust' to something like 'BBadust' or 'BBalpha' or 'BBalpha{dust}' (maybe) should work, as the texifying would then be done correctly. I expect you will also have problems with the parameters 'BBbetadust', 'BBalphasync' and 'BBbetasync', for the same reason.
Hi Thejs --
You mean that MontePython is reading a perfectly normal string out of log.param and silently turning it into a LaTeX-format label without any ability to configure this behavior? Not sure who thought that was a good idea. :-)
In any case, editing log.param to change "BBalphadust' to 'BBalphdust' etc. indeed worked. (Note as in my previous post, running with --extra did not, but failed with the error I describe above, which is a pity, because I note that there is a BK14.plot file that does the necessary parameter name conversions.)
It appears that MontePython --info correctly interprets $r$ as a single-tailed distribution, so the problem is with GetDist. See figure.
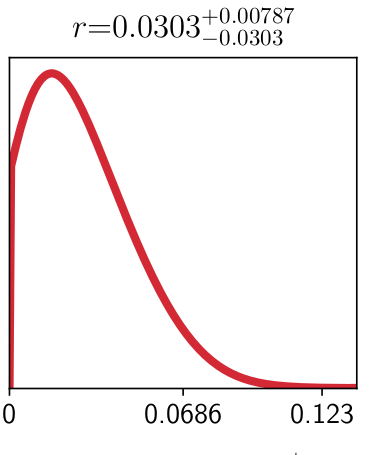
Ah I had misunderstood the problem with the plotting file. Issue #98 has a solution to the bug preventing you from passing a plot file. See the reply by Ivan Debono from January 24 (also yikes that's a long time ago, I really need to put out a bugfix asap!)
Great that the MontePython plotting is done correctly. I wonder how to provide GetDist with the necessary metadata files to analyze it correctly. Any ideas?
I'm guessing it has something to do with the .ranges file that is produced by cosmomc, but I can't seem to get that to work as of yet.
This is a nasty little bug, because GetDist doesn't fail, it just happily introduces O(10%) shifts in the parameter bounds...
Yikes. It should be easy to have MontePython construct a .ranges file for GetDist users. We'd definitely like for people to have an option of which analyze tool to use. I'll check it out and see if I can put something together.
Thanks for the help! I'll let you know if I come up with anything re: getting GetDist to work properly.
OK, interesting. I can fix the issue in GetDist with the setRanges() method in the MCSamples class, which converts $r$ to a correctly single-tailed distribution. Bounds are still not a good match though.

Have a look in my repository: https://github.com/ivandebono/montepython_utilities
The module mk_cosmomc_ranges in montepython2cosmomc.py will produce a .ranges file in the correct CosmoMC format from your log.param.
Thank you, Ivan! This is great! Would you like these included in the official release? I could create a new utilities directory and add them there, if you agree. In that case you should add the author blurb to montepython2cosmomc.py as well.
Best, Thejs
Yes, go ahead and include them. Feel free to modify the file structure if you wish to integrate the code in the main programme.