brainstorm3
 brainstorm3 copied to clipboard
brainstorm3 copied to clipboard
Global PCA for scouts and/or unconstrained sources
Description
This PR implements computing PCA across all files/epochs at once ("global pca") instead of separately for each file ("per-trial PCA"). It applies to extracting scout time-series, either through process_extract_scout or indirectly (e.g. for connectivity on scouts), and to "flattening" unconstrained source orientations to one source per vertex.
Justification
Following tests of various PCA procedures on the introductory tutorial data, it seems the feature of has the following advantages:
- It avoids sign flipping issues between trials. In particular, currently, PCA for unconstrained sources fails because of this, and the solution proposed by @rcassani for scouts does not work for unconstrained source flattening.
- It seems on par with per-trial PCA for extracting source activity across different conditions (e.g. many "standard" vs few "deviant" trials in the introduction tutorial), and should be more robust with very few or very short trials (though tests show per-trial is pretty robust too).
- It may simplify interpretation of comparisons when the same component is applied across conditions.
- It allows visualising the scout component maps.
- It can be used to do both scout and flattening in a single step, which is slightly better than doing flattening separately first, and noticeably better than doing scout extraction on x,y,z separately first. Here "better" means it keeps more source power, which is what PCA maximises. It achieves this by taking into account the full scout source covariance matrix, whereas doing it in 2 steps ignores some of the source pairs (e.g. location 1 x vs location 2 y).
Among these points, only 1 is pretty serious, but I think it all adds up to a good case for including this feature, and even using it as the default instead of per-trial PCA. A warning about the sign issue should also be added to per-trial PCA for flattening.
Implementation
This initial PR is a rough draft, how I implemented it for testing only. In particular, it has a bunch of extra options that would be removed (pcagt, pcag3, pcagx).
Currently, the functions that do PCA for scouts or flattening often see only one file at a time. Therefore, I implemented this using the data covariance matrix. The user would first compute the data covariance on all trials, and that would be used to compute the global PCA and applied to each file or to file averages, depending on the use case. Unfortunately, this means the same PCA decomposition is computed repeatedly each time it is applied to a file, but this was the simplest way to do it without creating new files (e.g. PCA kernels), or new fields (e.g. in the channel file, or inverse kernel file). And it is reasonably fast enough, equivalent to the current per-trial PCA.
I'm open to suggestions on how else we could implement this.
Documentation
Do we want to save somewhere a summary of the tests and results that show how the various PCA procedures perform? I have a couple slides now, but I could for example save it as a forum post which we could link to in the tutorials?
@ftadel indicated on the forum that he won't have time to review this before the fall.
Even though it's been like this for some time, I think the broken flattening PCA should be fixed asap.
Given this, I propose that I simplify this PR to only keep the global PCA option for flattening as a first step. Once reviewed and merged, I can introduce the rest of the changes for later review. Once the code is cleaned, I think it shouldn't be too long to review. Most of the changes are additions isolated in "if" blocks for the new option.
Even though it's been like this for some time, I think the broken flattening PCA should be fixed asap.
The PCA option is not used in any of our recommended pipelines, so it is not an emergency to do anything with it. All the things that are in the standard pipelines is much more urgent to fix (like the statistics for instance: https://github.com/brainstorm-tools/brainstorm3/issues/141, https://github.com/brainstorm-tools/brainstorm3/issues/225...)
Given this, I propose that I simplify this PR to only keep the global PCA option for flattening as a first step. Once reviewed and merged, I can introduce the rest of the changes for later review. Once the code is cleaned, I think it shouldn't be too long to review.
Sounds good.
Really trimmed down the changes to only unconstrained source flattening as a first step, for easier review. Only process_source_flat has access to the PCA function for orientations at the moment. This PR includes the sign fix of #534 .
Requested changes were made, so ready for another review once the "bugness" question is decided.
I added back flattening PCA per epochs, with sign correction based on PCA across epochs.
Some changes were made for efficiency, but it still takes quite a while to run flattening PCA per epochs: ~10 minutes for 192 epochs, vs seconds for across epochs which only runs once and saves a kernel for all files.
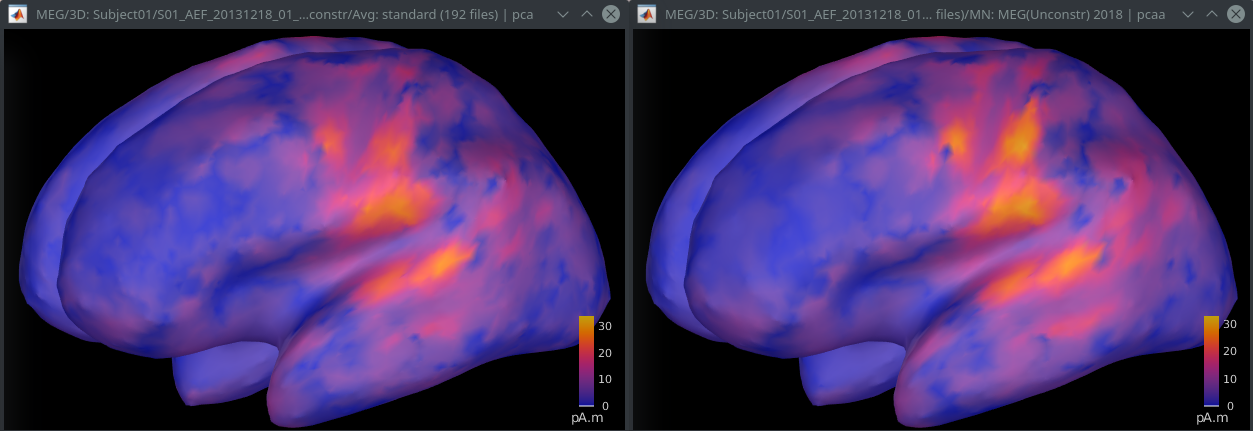
Tested, and confirmed similar results (left per epoch, right across epochs):
Thanks Francois for all the useful comments.
Regarding tutorials, how about a section for flattening right after the Unconstrained orientations section in the source estimation tutorial, or in the advanced section?
Regarding tutorials, how about a section for flattening right after the Unconstrained orientations section in the source estimation tutorial, or in the advanced section?
Maybe an advanced section just after: https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Averaging_normalized_values
Hi @ftadel, The code for PCA flattening is ready for review. It now works for non-kernel files, and across conditions/studies. (recall I kept non-kernel files for now since they'll be needed for scouts). If you can have a look to see how the flattening process is now organized and think a bit about scouts, I'd appreciate your input. I'm not sure yet what's the best approach since there are 2 extra levels of looping in process_extract_scout (scouts and frequencies).
There's now a new PCA options panel:
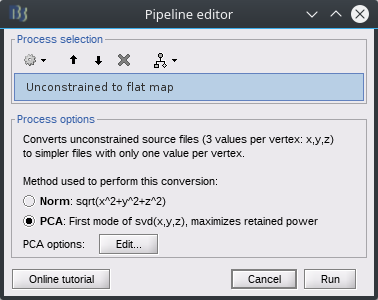
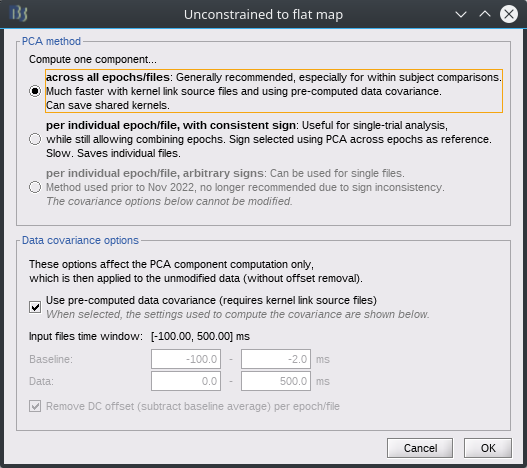
There's one useability bug I haven't yet figured out: when modifying options in the panel and clicking ok, it goes back to the process window. If we click the Edit button again, the panel options are reset, it doesn't keep the previous settings.
Note that I did pretty thorough tests of various inputs and settings. In particular, I confirmed the results are identical when using pre-computed data covariance or not, with the same settings.
(Commits incoming...) Here's the overview of changes in each file.
Main changes A toolbox/math/bst_pca.m
- Where the core of file operations now happen for doing PCA, except the final PCA computation which is still in bst_scout_value. It uses a lot of subfunctions from process_extract_scout. It is called by process_extract_scout, process_source_flat, and indirectly by all connectivity processes.
A toolbox/gui/panel_pca.m
M toolbox/process/functions/process_extract_scout.m
- Extensive code reorganization, modularized for reuse in bst_pca, but few functional changes
- PCA options
- New subfunctions for PCA scout extraction saved to temp files, and deleting those temp files, to be used by most processes that have scouts as an input option (or possibly in bst_process).
- Added compatibility checks for 'pca' across files: surface file, ncomp.
- change scoutfunc to radio_linelabel
- Output file .Comment used to be inconsistent for kernels vs full. Now "better", and added Method.
- .History now combines data file and inverse. (Inverse only has one line.)
M toolbox/math/bst_scout_value.m
- PCA, extra inputs
- stderr bug
- PCA scale change: each scout now more comparable to each other and to mean.
- (slightly stricter rules involving PCA, but doesn't restrict what was done by Bst previously)
- Fixed "explained", and prints separately for scout and flattening.
M toolbox/process/functions/process_source_flat.m
- Process type custom instead of file
- PCA options
- No longer save flattening method in Results.Function, which is inverse method.
M toolbox/inverse/bst_source_orient.m
- PCA, extra inputs: covariance and reference components;
- pcaa shortcut: projection on reference
M toolbox/gui/view_scouts.m
- PCA on scout & components at the same time instead of 3 x,y,z scouts (or norm)
M toolbox/core/bst_get.m
M toolbox/core/bst_set.m
- PcaOptions, typo "ndatacov"
Connectivity processes
M toolbox/process/functions/process_corr1n.m
M toolbox/process/functions/process_corr2.m
- PCA options
- change scoutfunc to radio_linelabel
- Fix single scout check for 1xN
M toolbox/process/functions/process_cohere1n_time_2021.m
- Fixed time window bug
M toolbox/process/functions/process_cohere1_2021.m
M toolbox/process/functions/process_cohere1n_2021.m
M toolbox/process/functions/process_cohere2_2021.m
M toolbox/process/functions/process_cohere2_time_2021.m
M toolbox/process/functions/process_corr1.m
M toolbox/process/functions/process_corr1n_time.m
M toolbox/process/functions/process_granger1.m
M toolbox/process/functions/process_granger1n.m
M toolbox/process/functions/process_granger2.m
M toolbox/process/functions/process_henv1.m
M toolbox/process/functions/process_henv1n.m
M toolbox/process/functions/process_henv2.m
M toolbox/process/functions/process_plv1.m
M toolbox/process/functions/process_plv1n.m
M toolbox/process/functions/process_plv2.m
M toolbox/process/functions/process_pte1n.m
M toolbox/process/functions/process_spgranger1.m
M toolbox/process/functions/process_spgranger1n.m
M toolbox/process/functions/process_spgranger2.m
- Call PCA for scouts first, delete temp files after
- For 1xN: keep copy of non-scout files for "B" inputs.
Other minor non-PCA specific changes
M toolbox/tree/tree_view_scouts.m
- Improved behaviour for displaying scouts from tree
M toolbox/connectivity/bst_connectivity.m
- Save applied ScoutFunction, fix AxB output file comment.
M toolbox/math/bst_noisecov.m
- (no longer needed by PCA – could be reverted) isSave option and hide progress for 1 file.
M toolbox/gui/panel_time.m
M toolbox/io/in_bst.m
M toolbox/process/bst_process.m
- (comment only)
Finally the PR is ready for review again @rcassani.
Scout and flattening PCA are implemented and all connectivity processes updated. Most of the file manipulation for PCA across files is now done in a new bst_pca function.
I did pretty thorough testing with many different combinations of input files: different conditions, types of result files (links, kernels, full), constrained/unconstrained/loose/mixed, etc., with process_extract_scout and process_source_flat. For connectivity, all 22 non-deprecated processes were tested with one set of options each, varying options among them, but always using scouts.
Still to do:
- Documentation.
- Possibly: change bst_process to do the temporary scout extraction instead of each process.
- Time-frequency processes that use scouts.
Ready for final review & merging.
Recent changes were mainly to reproduce exactly the previous PCA behavior with the "legacy" (grayed out) option. However, "old script reproducibility" is not maintained. This topic was discussed and a new tutorial page will be dedicated to it, to make it easier for users to get older versions of Brainstorm. For this PR, new recommended options are the defaults and old scripts that are missing the new options would run with defaults or saved user preferences.
On the other hand, time frequency processes (incl. PAC) still need to be updated to use the new PCA, in a separate PR, and until then they will still be using the deprecated legacy PCA. Similarly for PCA on clusters of sensors.