Implement mechanism for supporting different views of the same information
Different use cases require different views or perspectives on the data model. We should implement a computable system for declaring how to transform between views. Some of these will be information preserving, some will be lossy
For example, for modeling a metabolic pathway, we can imagine at least 3 views:
- substance-centric graph, with one node per substance, and edges denoting transitions (excluding currency chemicals). E.g.
chem1->chem2->... - gene-centric, where we model the sequence of gene activities needed to complete the metabolic process. E.g.
g1->g2->... - Granular model with a bipartite graph of reactions and entities, with reactions connected to entities via has-catalyst/substrate/product
Another example is a reactome-style pathway model to go-cam:

Another example is going from an activity-centric (e.g. GO-CAM view) to a simple entity-entity view:
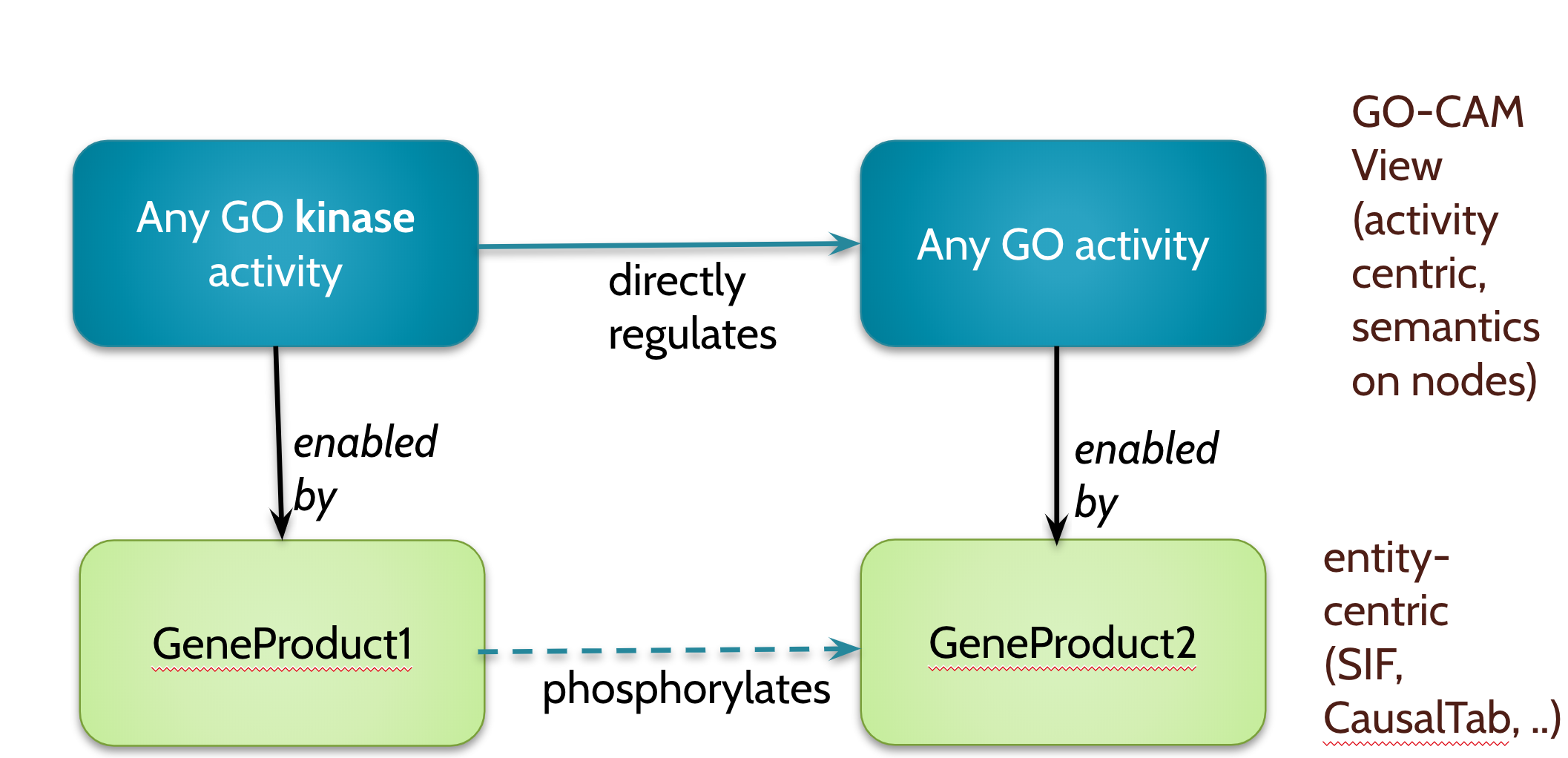
The BioLink Model should define these transforms. This is non-trivial since this is a transform not a mapping. We want to keep it declarative and avoid complex hard to interpret code. Something like SWRL may work here. Or kanren-style rules (cc @webyrd)
@cmungall @deepakunni3 Can you briefly comment on the status of this? Thanks!
The idea of different views for Biolink Model has not yet been explored in detail. Would need a bit more clarification from @cmungall on the priority of this task.
@deepakunni3, @cmungall,
I wonder if standards like Systems Biology Graphical Notation (SBGN; https://sbgn.github.io/) might help us wrap our heads around what is needed here? Sometime during the NCATS Feasibility phase, I had pondered the idea of somehow computationally cross-linking certain Translator subgraphs (i.e. that involve relevant Biolink concept categories and relationship predicates) to some kind of SBGN description.
SBGN is a bit molecular biology centric. Maybe for other kinds of biomedical knowledge, other diagramatical formalisms might be available to tap into.
@cmungall Is the transform 1-to-1 and onto?
@cmungall this reminds me of bidirectional programming: https://en.wikipedia.org/wiki/Bidirectional_transformation https://www.cis.upenn.edu/~bcpierce/papers/icmt-2009-slides.pdf
This is the functional-programming equivalent of the "view-update" problem in databases.
What is the status of this?
@cmungall - do you think this is still a relevant idea for us to work towards?
@tursynay asked whether we're moving ahead with this.