Gff fails to parse on basic Gff file
package main
import (
"fmt"
"github.com/koeng101/poly"
)
// Started at 4:44
// 5:00 finished downloading all files
// 5:07 discovered big bug with gb file parsing (give this to poly folks)
func main() {
// Parse Genbank file
//ct := poly.GetCodonTable(11)
sequence := poly.ReadGff("data/Arthrospira_platensis.gff")
for _, feature := range sequence.Features {
if feature.Type == "CDS" {
fmt.Println(feature)
}
}
// Find associated uniprot numbers
// Find associated Rhea reaction numbers
// Find associated Rhea
}
Like #152 , koeng101/poly has an up-to-date io.go file.
Output
panic: runtime error: index out of range [2] with length 2
goroutine 1 [running]:
github.com/koeng101/poly.ParseGff(0xc000180000, 0x31b6e7, 0x31b6e8, 0x0, 0x0, 0x0, 0x0, 0x0, 0x0, 0x0, ...)
/home/koeng/go/pkg/mod/github.com/koeng101/[email protected]/io.go:160 +0xaf9
github.com/koeng101/poly.ReadGff(0x645b01, 0x1e, 0x0, 0x0, 0x0, 0x0, 0x0, 0x0, 0x0, 0x0, ...)
/home/koeng/go/pkg/mod/github.com/koeng101/[email protected]/io.go:333 +0xb4
main.main()
/home/koeng/go/src/github.com/koeng101/fun_spira_project/main.go:15 +0x4b
exit status 2
We should be able to parse this file. Arthrospira_platensis.gff.gz
And here I thought we'd never actually have someone use the gff parser.
@Koeng101 has this been solved yet?
No don't think so. I think I actually just solved the problem that was preventing me from using the raw genbank (which forced me to use gff). Maybe we should depreciate GFF for a while until we can get proper error handling on it? Or keep it, but note it is buggy or whatever.
I think I have an idea how to go about it....
Problem
-
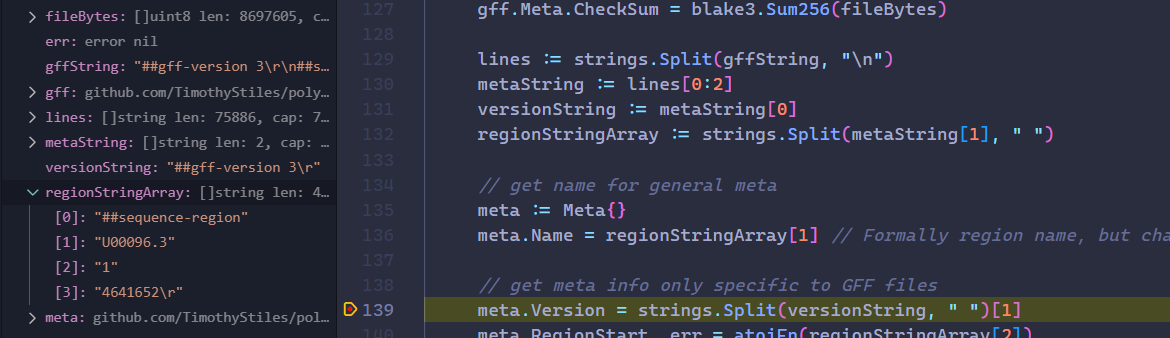
here this is a snippet from gff.go, for the
./data/ecoli-mg1655.gff, in line 132 it is assumed that the second line is always a place for##sequence-region -
-
the file
Arthrospira_platensis.gffhas##sequence-regionon line 8,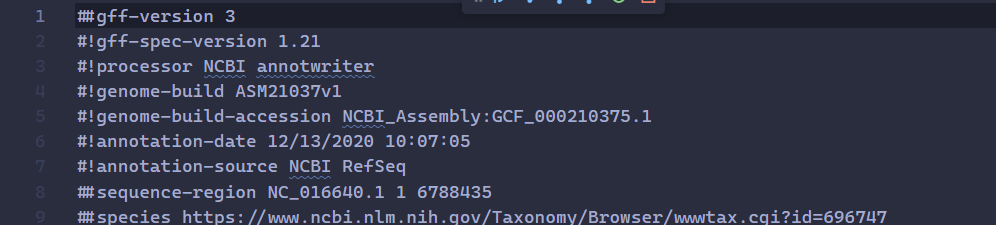
- on line 2,
#!gff-spec-version 1.21has only two items in the split array, hence we get the index out of bounds error
-
thing is this specific gff file has more than 2 line of meta, which is causing the parser to fail