Add NEON implementation for `armv7` and `aarch64` platforms
Hi Benjamin!
Overview
As mentioned on Twitter I managed to get a working port of the vectorized code to Arm using NEON extensions. Most of the code is compatible with both armv7 and aarch64 platforms, with the exception of a few table-lookup based functions that are only available for aarch64.
I added NEON implementations for score vectors, SSE dist, fingerprints, and 16x16 matrix transpose, which I think covers pretty much everything for which there is SSE support. In addition, I also added a faster BitVector::one_count, since there is a vectorized population count instruction available in NEON.
Todo
Some things left for me to do:
- [x] Add runtime detection of NEON for
armv7(onaarch64NEON is mandatory). ~~For this I'll likely needgetauxval~~. Done:getauxvalis detected by CMake, and if available is then used to set the flags insimd.cpp. - [x] Add compile-time detection of NEON (currently all Arm builds will attempt to build with
-mfpu=neon, which will break on older platforms without NEON support). Done:arch_neonand the NEON dispatch mechanism will be build inconditionally onaarch64but only when-mfpu=neonis supported onarmv7. - [ ] Fix traceback issues on
armv7or droparmv7support.
Tests
I don't have access to a Mac to test it. If you want, I can also setup a GitHub Action to compile on aarch64, but it tend to be quite slow because there are no native runners available so it's necessary to use QEMU.
So far everything compiles and works on my Raspberry Pi 4 (aarch64):
$ uname -a
Linux chloroplast 5.10.0-17-arm64 #1 SMP Debian 5.10.136-1 (2022-08-13) aarch64 GNU/Linux
$ gcc --version
gcc (Debian 10.2.1-6) 10.2.1 20210110
$ ./diamond test
diamond v2.0.15.153 (C) Max Planck Society for the Advancement of Science
Documentation, support and updates available at http://www.diamondsearch.org
Please cite: http://dx.doi.org/10.1038/s41592-021-01101-x Nature Methods (2021)
blastp (default) [ Passed ]
blastp (multithreaded) [ Passed ]
blastp (blocked) [ Passed ]
blastp (more-sensitive) [ Passed ]
blastp (very-sensitive) [ Passed ]
blastp (ultra-sensitive) [ Passed ]
blastp (max-hsps) [ Passed ]
blastp (target-parallel) [ Passed ]
blastp (query-indexed) [ Passed ]
blastp (comp-based-stats 0) [ Passed ]
blastp (comp-based-stats 2) [ Passed ]
blastp (comp-based-stats 3) [ Passed ]
blastp (comp-based-stats 4) [ Passed ]
blastp (target seqs) [ Passed ]
blastp (top) [ Passed ]
blastp (evalue) [ Passed ]
blastp (blosum50) [ Passed ]
blastp (pairwise format) [ Passed ]
blastp (XML format) [ Passed ]
blastp (PAF format) [ Passed ]
#Test cases passed: 20/20
$ ./diamond benchmark
diamond v2.0.15.153 (C) Max Planck Society for the Advancement of Science
Documentation, support and updates available at http://www.diamondsearch.org
Please cite: http://dx.doi.org/10.1038/s41592-021-01101-x Nature Methods (2021)
Scoring parameters: (Matrix=BLOSUM62 Lambda=0.267 K=0.041 Penalties=11/1)
SWIPE (int8_t): 5574.8 ps/Cell
SWIPE (int8_t, Stats): 5574.53 ps/Cell
SWIPE (int8_t, MatrixAdjust): 5638.35 ps/Cell
SWIPE (int8_t, CBS): 5638.21 ps/Cell
SWIPE (int8_t, TB): 5638.21 ps/Cell
Diagonal scores: 348.589 ps/Cell
Banded SWIPE (int16_t, CBS): 2888.76 ps/Cell
Banded SWIPE (int16_t): 2889.27 ps/Cell
Banded SWIPE (int16_t, CBS, TB):2888.82 ps/Cell
Evalue: 131.707 ns
Evalue (ALP): 844.882 ns
Matrix adjust: 1036.13 ms
Matrix adjust (vectorized): 692.898 micros
NEON hamming distance: 324.783 ps/Cell
Scalar ungapped extension: 10309.4 ps/Cell
NEON score shuffle: 730.79 ps/Letter
Transpose (16x16): 4711.21 ps/Letter
Transpose (16x16, vectorized): 469.893 ps/Letter
On a Beaglebone Black (armv7), code compiles, but tests fail at the moment, I need to look into:
$ uname -a
Linux beaglebone 4.19.94-ti-r42 #1buster SMP PREEMPT Tue Mar 31 19:38:29 UTC 2020 armv7l GNU/Linux
$ gcc --version
gcc (Debian 8.3.0-6) 8.3.0
$ ./diamond test
diamond v2.0.15.153 (C) Max Planck Society for the Advancement of Science
Documentation, support and updates available at http://www.diamondsearch.org
Please cite: http://dx.doi.org/10.1038/s41592-021-01101-x Nature Methods (2021)
Error: Traceback error.
$ ./diamond benchmark
diamond v2.0.15.153 (C) Max Planck Society for the Advancement of Science
Documentation, support and updates available at http://www.diamondsearch.org
Please cite: http://dx.doi.org/10.1038/s41592-021-01101-x Nature Methods (2021)
Scoring parameters: (Matrix=BLOSUM62 Lambda=0.267 K=0.041 Penalties=11/1)
SWIPE (int8_t): 6923.98 ps/Cell
SWIPE (int8_t, Stats): 6922.9 ps/Cell
SWIPE (int8_t, MatrixAdjust): 7077.76 ps/Cell
SWIPE (int8_t, CBS): 7076.77 ps/Cell
SWIPE (int8_t, TB): 7080.45 ps/Cell
Diagonal scores: 388.169 ps/Cell
Banded SWIPE (int16_t, CBS): 5398.94 ps/Cell
Banded SWIPE (int16_t): 5400.96 ps/Cell
Banded SWIPE (int16_t, CBS, TB):5400.45 ps/Cell
Evalue: 1398.97 ns
Evalue (ALP): 6293.59 ns
Matrix adjust: 6631.94 ms
Matrix adjust (vectorized): 5295.68 micros
NEON hamming distance: 9783.43 ps/Cell
Scalar ungapped extension: 14621.5 ps/Cell
Transpose (16x16): 8811.47 ps/Letter
Transpose (16x16, vectorized): 2188.78 ps/Letter
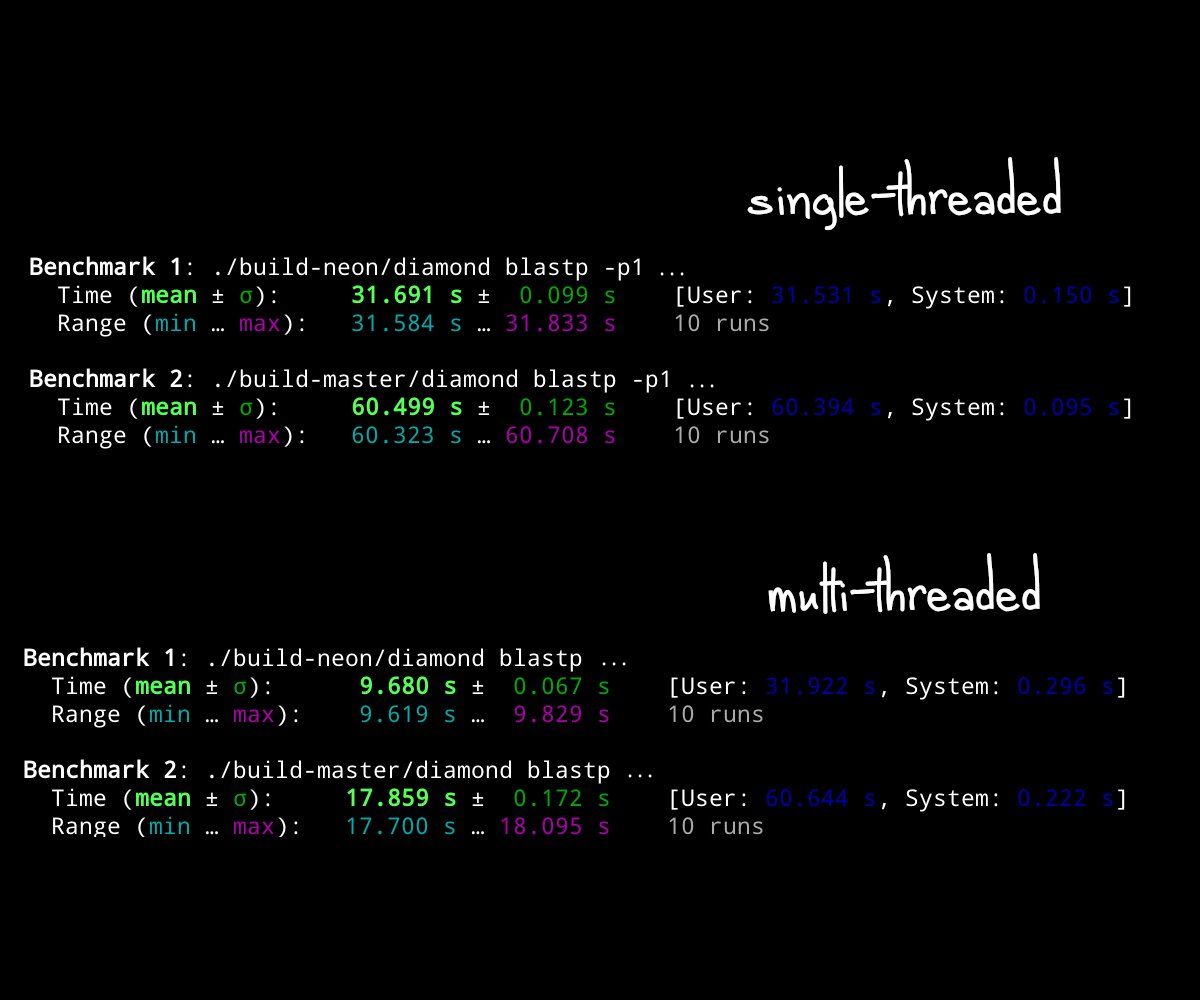
Benchmarks
On a Raspberry Pi 4:
Hi Martin. Thanks, that's really great work and very happy that it will finally be possible to run Diamond on ARM with vectorization. Please permit me some time to look this over and integrate it into the next release.
No problem, I know this is a lot of code to take in, so I'm not expecting an immediate merge. I'm happy to answer any questions you may have :)
Sorry that I left this. Unfortunately, my private development branch had already diverged a lot from the public master when you posted this, so it wasn't possible to easily merge this, and still isn't. I still would like to merge it though. Please let me know if you are interested in resolving the merge conflicts. Otherwise I will put it on my todo list or we may be able to find a student to do it.
Most of it should be resolved (with the master branch at least), I'm checking on an armv7l platform. Are there any parts that have been updated with new SIMD code? I have yet to enable SIMD in pfscan.cpp but I'm wondering if there are other places for me to look at.
@bbuchfink : Done fixing the merge conflicts! In addition I also fixed the armv7 impl so now the tests pass on there as well (the ones i can run on my limited memory platform, cause at some point i'm getting an error that I can't allocate more memory).
Great thanks, I quickly tested this and everything seems to be working. Should be no problem to have this merged for the next release.