feat(bayesian_optimization.py): Allow registering of temporary observations
Potential fix for #347
Codecov Report
Base: 99.25% // Head: 97.16% // Decreases project coverage by -2.08% :warning:
Coverage data is based on head (
7547acc) compared to base (8e5c89f). Patch coverage: 85.29% of modified lines in pull request are covered.
Additional details and impacted files
@@ Coverage Diff @@
## master #365 +/- ##
==========================================
- Coverage 99.25% 97.16% -2.09%
==========================================
Files 8 8
Lines 535 564 +29
Branches 81 89 +8
==========================================
+ Hits 531 548 +17
- Misses 2 13 +11
- Partials 2 3 +1
| Impacted Files | Coverage Δ | |
|---|---|---|
| bayes_opt/target_space.py | 92.41% <71.42%> (-7.59%) |
:arrow_down: |
| bayes_opt/bayesian_optimization.py | 99.16% <95.00%> (-0.84%) |
:arrow_down: |
Help us with your feedback. Take ten seconds to tell us how you rate us. Have a feature suggestion? Share it here.
:umbrella: View full report at Codecov.
:loudspeaker: Do you have feedback about the report comment? Let us know in this issue.
Hey @Rizhiy, thanks for contributing!
I (superficially) follwed the discussion in the original issue and I agree with the notions that a) this package should be compatible with parallel optimization and b) in it's current state, that can't really be considered the state (as you have shown). Nonetheless, I'm always somewhat sceptical of messing with internal data and even moreso here, since it involves providing "wrong" values. Is there some way that this could go wrong? Off the top of my head:
- What happens if there are duplicate points (i.e. points with the same parameters), would it be possible that the "real" point is updated instead of the dummy?
- What happens if the optimization process terminates early (maybe some stopping criterion is reached), will dummies persist?
If I understand correctly, the idea behind this technique of registering dummies is to reduce the covariance of the GPR at a point that is being sampled so that other workers are suggested other points. Is there a way to perform this covariance reduction without registering dummies? Alternatively, is there a way to force different points to be returned without using dummies, e.g.
- ...by changing the
pbounds? - ...messing with
x_seedsinacq_maxso that only seeds are used which are a certain distance away from the last suggestion? - ...penalizing points in the objective
to_minimizeofacq_maxthat are close to points other workers are considering? (this is probably what I would try)
In particular, I think that modifying acq_max seems to be a more stable/more elegant way of achieving what you're going for: To my understanding, what you're trying to accomplish is for the optimizer to suggest points that are away from points currently investigated by other workers. The acq_max function is the place where suggestions are made, so I would also expect it to be the appropriate place for your modifications.
Hi @till-m, regarding your concerns:
What happens if there are duplicate points (i.e. points with the same parameters), would it be possible that the "real" point is updated instead of the dummy?
Not sure what you mean, dummy points are stored in _dummies variable:
https://github.com/Rizhiy/BayesianOptimization/blob/94cd1a29ab12e334f1f60528a03cea1642f82786/bayes_opt/bayesian_optimization.py#L135
Interactions for other observations should be the same, can you perhaps try it out and I can correct the wrong behaviour.
What happens if the optimization process terminates early (maybe some stopping criterion is reached), will dummies persist?
They persist until you overwrite them.
I would guess to make it more user-friendly, the relevant functions (e.g. max & res) should include the filter for dummies.
Is there a way to perform this covariance reduction without registering dummies?
There very well might be, but I haven't dived deep into how the optimiser works on the inside yet. If you know a better way, please let me know and I will update this Pull Request. For my approach I only need the functionality to let the optimiser know that a point is being evaluated.
In particular, I think that modifying acq_max seems to be a more stable/more elegant way of achieving what you're going for
I will look into that.
Hi @Rizhiy,
I misread the code and after thinking about it again, I think you've covered the scenario that I was picturing. My apologies.
@till-m I have looked into other ways you suggested achieving similar effect and currently it seems that they won't be as effective or too difficult to implement. Basically, it is very difficult to properly define which area exactly we don't want to sample.
There very well might be a better approach for parallel evaluation, but I haven't been able to find any yet.
Looking more closely at the code, perhaps it would make more sense to put dummy logic into TargetSpace class, since it has kind of similar logic with constraints.
Hi @Rizhiy,
I would guess to make it more user-friendly, the relevant functions (e.g. max & res) should include the filter for dummies.
I agree. I think logging should also be considered -- dummies probably shouldn't show up in logs, especially when the JSONLogger is used, so that loading from logs doesn't register the dummies normally.
Looking more closely at the code, perhaps it would make more sense to put dummy logic into TargetSpace class, since it has kind of similar logic with constraints.
Agreed.
I have looked into other ways you suggested achieving similar effect and currently it seems that they won't be as effective or too difficult to implement.
Thanks for looking into this. I also had a look around the literature and I found a few potentially interesting things. Your approach seems to be related to "Constant Liars", see Ginsbourger et al; 2008. Consider also the "Kriging Believer" which may be a more natural way to pick the values of the dummies.
I would also like to highlight the approach outlined in Batch Bayesian Optimization via Local Penalization (see also here), that works by penalizing the acquisition function and would be close to what I was thinking of. Finally, Kandasamy et al might also be worth considering, since it seems to work very well while not being too hard to implement, at least at first glance.
There's a lot more literature on this problem than I thought there would be, two others that I found potentially interesting would be:
- Wang et al; Parallel Bayesian Global Optimization of Expensive Functions, 2016
- Snoek et al; Practical Bayesian Optimization of Machine Learning Algorithms, 2012
Hi @Rizhiy, I decided to write a quick implementation of Kandasamy to see it work. The preliminary results are pretty disappointing, will post later.
Okay, so first of all it looks like sampling from the multivariate normal is very computationally expensive. I had to adapt n_warmup in acq_max to make this feasible, so I'm now using 1e4 instead of 1e5. I've adapted your code from the issue.
Here are the results:
Output:
Checking num_workers: 100%|██████████████████████| 4/4 [12:11<00:00, 182.88s/it]
num_workers=1, mean_min_distance=0.406, best_result=-0.171
num_workers=2, mean_min_distance=0.468, best_result=-0.276
num_workers=4, mean_min_distance=0.359, best_result=-0.707
num_workers=8, mean_min_distance=0.419, best_result=-0.112
Plot:
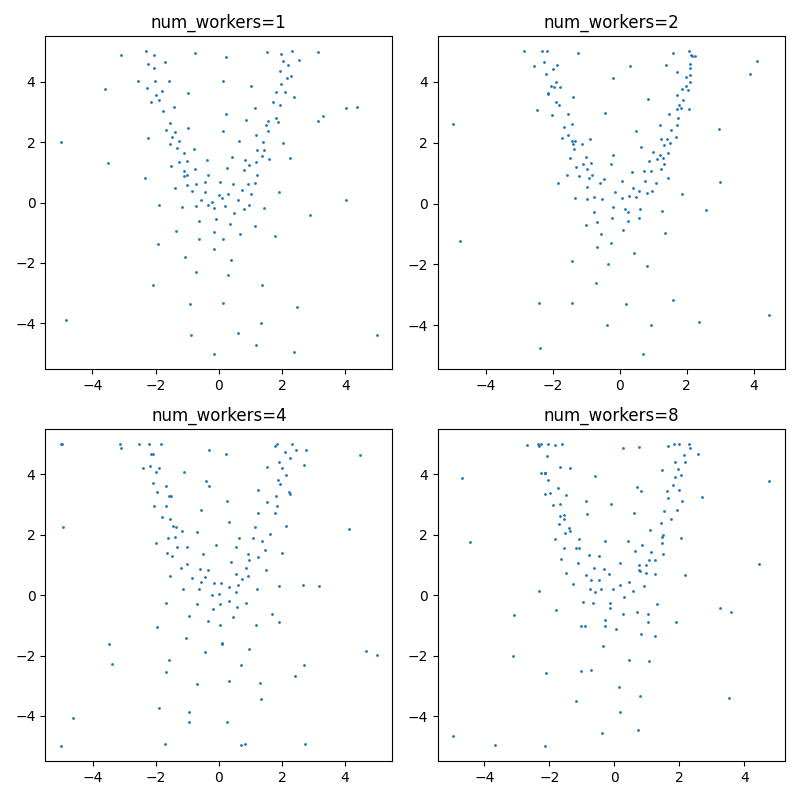
I'm somewhat disappointed with these results. It doesn't really seem to hone in on the max very well. Additionally, sampling from the multivariate normal also seems to be quite computationally intensive -- I takes around 4x as long to run the script with async_thompson=True. Admittedly my optimization loop is very naive, but I currently don't see a way of speeding this up.
Output (async_thompson=False)
Checking num_workers: 100%|███████████████████████| 4/4 [03:18<00:00, 49.56s/it]
num_workers=1, mean_min_distance=0.634, best_result=-3.440
num_workers=2, mean_min_distance=0.320, best_result=-14.352
num_workers=4, mean_min_distance=0.192, best_result=-3.855
num_workers=8, mean_min_distance=0.209, best_result=-1.974
from __future__ import annotations
from typing import Callable
import matplotlib.pyplot as plt
import numpy as np
from bayes_opt.bayesian_optimization import BayesianOptimization
from bayes_opt.util import UtilityFunction
from pandas.core.generic import pickle
from scipy.optimize import rosen
from scipy.stats import multivariate_normal
from tqdm import tqdm
from sklearn.gaussian_process import GaussianProcessRegressor
import warnings
class asyTS():
def __init__(self) -> None:
self.random_state_generator = lambda: np.random.randint(0, 100000)
@property # I know that this is giga-hacky monkey patching, please bear with me.
def utility(self):
random_state = self.random_state_generator()
def func(x, gp: GaussianProcessRegressor, y_max):
with warnings.catch_warnings():
warnings.simplefilter("ignore")
# fixing random state causes it to return the same function
return gp.sample_y(x, random_state=random_state)
return func
def _closest_distance(point, points):
return min(np.linalg.norm(point - p) for p in points if p is not point)
def optimize(
func: Callable[..., float], num_iter: int, bounds: dict[str, tuple[float, float]], num_workers=0, async_thompson=True
):
init_samples = int(np.sqrt(num_iter))
optimizer = BayesianOptimization(f=None, pbounds=bounds, verbose=0)
init_kappa = 10
kappa_decay = (0.1 / init_kappa) ** (1 / (num_iter - init_samples))
if async_thompson:
utility = asyTS()
else:
utility = UtilityFunction(
kind="ucb", kappa=init_kappa, xi=0.0, kappa_decay=kappa_decay, kappa_decay_delay=init_samples
)
init_queue = [optimizer.suggest(utility) for _ in range(init_samples)]
result_queue = []
tbar = tqdm(total=num_iter, leave=False)
while len(optimizer.res) < num_iter:
sample = init_queue.pop(0) if init_queue else optimizer.suggest(utility)
loss = func(list(sample.values())) * -1
result_queue.append((sample, loss))
if len(result_queue) >= num_workers:
try:
optimizer.register(*result_queue.pop(0))
tbar.update()
except KeyError:
pass
return optimizer.res
bounds = {"x": [-5, 5], "y": [-5, 5]}
all_results = {}
for num_workers in tqdm([1, 2, 4, 8], desc="Checking num_workers"):
results = []
results = optimize(rosen, 150, bounds, num_workers, async_thompson=True)
all_results[num_workers] = results
with open("results.pkl", "wb") as f:
pickle.dump(all_results, f)
with open("results.pkl", "rb") as f:
all_results = pickle.load(f)
fig, axs = plt.subplots(2, 2)
fig.set_figheight(8)
fig.set_figwidth(8)
axs = [item for sublist in axs for item in sublist]
for idx, (num_workers, results) in enumerate(all_results.items()):
if num_workers > 8:
continue
samples = [np.array(list(res["params"].values())) for res in results]
axs[idx].scatter(*zip(*samples), s=1)
axs[idx].set_title(f"{num_workers=}")
avg_min_distance = np.mean([_closest_distance(sample, samples) for sample in samples])
best_result = max([res["target"] for res in results])
print(f"{num_workers=}, mean_min_distance={avg_min_distance:.3f}, {best_result=:.3f}")
fig.tight_layout()
fig.savefig("results.png")
Is there something similar to kappa decay in your code?
For the algorithm to hone in on the maximum better you need to progressively reduce kappa to 0.
In my code there is utility.update_params() which is called after every register, it multiplies kappa by kappa_decay which makes the model less explorative and more exploitative.
My apologies for the delay.
Is there something similar to kappa decay in your code?
In theory this shouldn't be necessary, but I could add it. I will try to get around to doing that. Overall, I'm not sure if the TS approach is worth exploring due to the sampling speed.
Hi @Rizhiy,
apologies for letting this sit for a while. I've been thinking and we should definitely have functionality for this, but it would be better to first redesign the acquisition function API. I've implemented some ideas and will push them shortly, including an implementation of the Kriging believer algorithm. Would you be interested in reviewing and/or testing that code?
Hi, I can go over it briefly, but TBH I haven't been looking in this area since last year, since my approach works fine for me.
Closing as this is handled in #447. @Rizhiy I adapted your benchmarking code as an example incl. attribution. I hope that's okay.