panel-chemistry
 panel-chemistry copied to clipboard
panel-chemistry copied to clipboard
🧪📈 🐍. The purpose of the panel-chemistry project is to make it really easy for you to do DATA ANALYSIS and build powerful DATA AND VIZ APPLICATIONS within the domain of Chemistry using using Pytho...
🧪 Panel-Chemistry
👨🔬📈🛠️🐍❤️
The purpose of the panel-chemistry project is to make it really easy for you to do exploratory data analysis and build powerful data and viz tools within the domain of Chemistry using Python and HoloViz Panel.

Check out the panel-chemistry examples on Binder
| Jupyter Notebook | Jupyter Labs | Panel Apps |
|---|---|---|
🏁 Background
This project was started by the discussion How to display JSME molecular editor with Panel? in the Panel Community Forum
🏃 Getting Started
pip install panel-chemistry
or with conda
conda install -c conda-forge panel-chemistry
Run the below in a Notebook or using panel serve NAME_OF_SCRIPT.py.
import panel as pn
from panel_chemistry.widgets import JSMEEditor
pn.extension("jsme", sizing_mode="stretch_width")
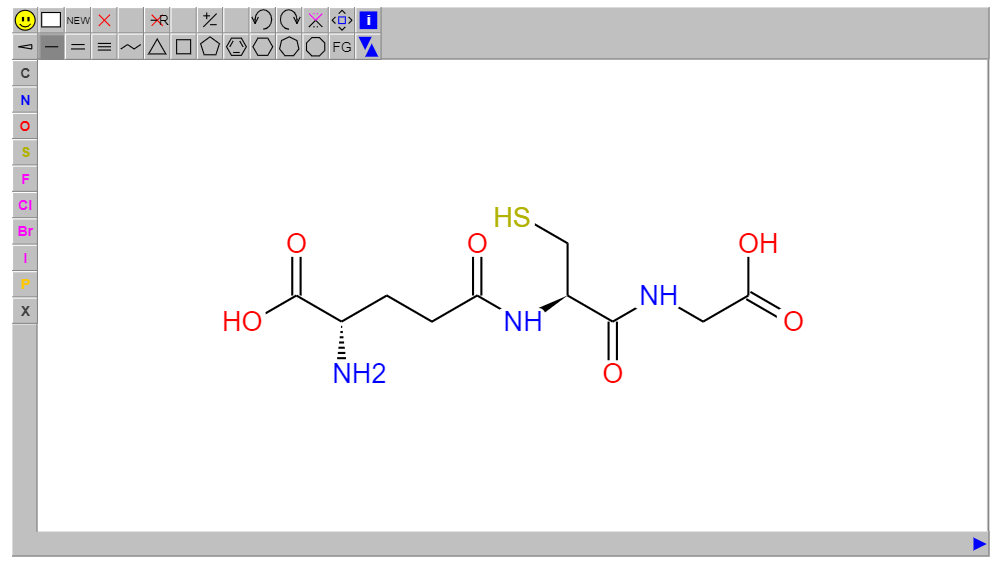
smiles="N[C@@H](CCC(=O)N[C@@H](CS)C(=O)NCC(=O)O)C(=O)O"
editor = JSMEEditor(value=smiles, height=500, format="smiles")
editor.servable()
👩🏫 Examples
Check out the panel-chemistry reference guides on Binder
| Guide | NB Viewer | Github Notebook | Jupyter Notebook | Jupyter Labs | Panel Apps |
|---|---|---|---|---|---|
| JSME Editor | View | View | |||
| NGL Viewer | View | View | |||
| PDBe_MolStar | View | View | |||
| Py3DMol Pane | View | View |
💡 Inspiration
- Awesome Python Chemistry
- Dash Bio
- JSME Editor and Test Page
- 3DMol.js and py3Dmol
- RDKit
- RDKit IPython Tools
🎁 Contributing
If you want to contribute reach out via Github Issues or in the Contributor Community Forum on Gitter: https://gitter.im/panel-chemistry/community#
For more details see the Developer Guide
FAQ
📰 Change Log
- 0.0.12: Add PDBeMolstar component and py.typed file
- 0.0.11: Add LICENSE and VERSION files to package. Now available on conda-forge
- 0.0.10: Update to Panel 0.12.6.
- 0.0.9: Add Py3DMol pane. Update to Panel 0.12.4.


