Mapping Difference
Hi,
I am trying to map atoms of molecules supported by Recon 3D. In Recon 3D study Reaction Decoder tool have been also used, and the mapped reactions can be found in Virtual Metabolic Human.
Here is my problem. When i performed AAM in the same reactions by using SMILES of metabolites which are obtained from VHM, i can not obtained the same mapping images.
As an example: DHCR72r reaction in cholestrol metabolism
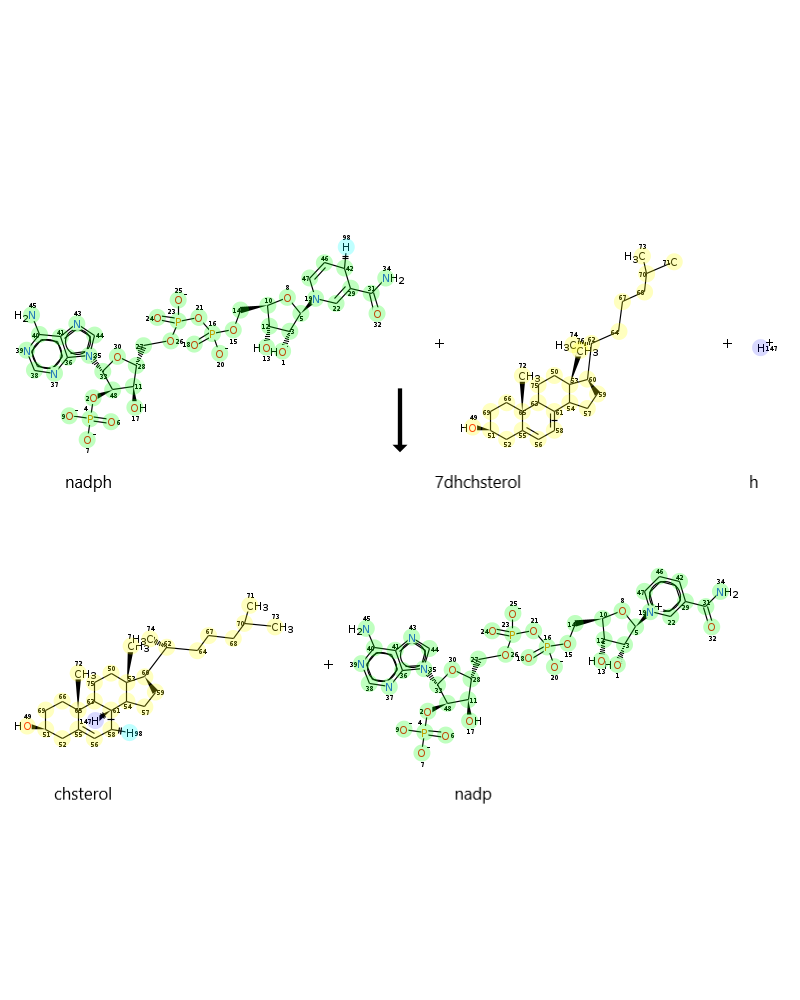
When it was searched DHCR72r reaction in VMH, following image are found:

-
Reaction is given as 7dhchsterol[r] + h[r] + nadph[r] -> chsterol[r] + nadp[r]
-
Metabolites are
7dhchsterol:
[H]O[C@]1([H])C([H])([H])C2=C([H])C([H])=C3[C@]4([H])C([H])([H])C([H])([H])[C@]([H])([C@]([H])(C([H])([H])[H])C([H])([H])C([H])([H])C([H])([H])C([H])(C([H])([H])[H])C([H])([H])[H])[C@@]4(C([H])([H])[H])C([H])([H])C([H])([H])[C@]3([H])[C@@]2(C([H])([H])[H])C([H])([H])C1([H])[H]
h:
[H+]
nadph:
[H]O[C@@]1([H])[C@@]([H])(O[C@]([H])(C([H])([H])OP([O-])(=O)OP([O-])(=O)OC([H])([H])[C@@]2([H])O[C@@]([H])(N3C([H])=NC4=C(N=C([H])N=C34)N([H])[H])[C@]([H])(OP([O-])([O-])=O)[C@]2([H])O[H])[C@@]1([H])O[H])N1C([H])=C([H])C([H])([H])C(=C1[H])C(=O)N([H])[H]
products:
chsterol:
[H]O[C@]1([H])C([H])([H])C2=C([H])C([H])([H])[C@@]3([H])[C@]4([H])C([H])([H])C([H])([H])[C@]([H])([C@]([H])(C([H])([H])[H])C([H])([H])C([H])([H])C([H])([H])C([H])(C([H])([H])[H])C([H])([H])[H])[C@@]4(C([H])([H])[H])C([H])([H])C([H])([H])[C@]3([H])[C@@]2(C([H])([H])[H])C([H])([H])C1([H])[H]
nadp:
[H]O[C@]1([H])[C@]([H])(O[H])[C@]([H])(O[C@@]1([H])[N+]1=C([H])C(=C([H])C([H])=C1[H])C(=O)N([H])[H])C([H])([H])OP([O-])(=O)OP([O-])(=O)OC([H])([H])[C@@]1([H])O[C@@]([H])(N2C([H])=NC3=C(N=C([H])N=C23)N([H])[H])[C@]([H])(OP([O-])([O-])=O)[C@]1([H])O[H]
- So, reaction input is constructed as:
rxn:
[H]O[C@]1([H])C([H])([H])C2=C([H])C([H])=C3[C@]4([H])C([H])([H])C([H])([H])[C@]([H])([C@]([H])(C([H])([H])[H])C([H])([H])C([H])([H])C([H])([H])C([H])(C([H])([H])[H])C([H])([H])[H])[C@@]4(C([H])([H])[H])C([H])([H])C([H])([H])[C@]3([H])[C@@]2(C([H])([H])[H])C([H])([H])C1([H])[H].[H+].[H]O[C@@]1([H])[C@@]([H])(O[C@]([H])(C([H])([H])OP([O-])(=O)OP([O-])(=O)OC([H])([H])[C@@]2([H])O[C@@]([H])(N3C([H])=NC4=C(N=C([H])N=C34)N([H])[H])[C@]([H])(OP([O-])([O-])=O)[C@]2([H])O[H])[C@@]1([H])O[H])N1C([H])=C([H])C([H])([H])C(=C1[H])C(=O)N([H])[H]>>[H]O[C@]1([H])C([H])([H])C2=C([H])C([H])([H])[C@@]3([H])[C@]4([H])C([H])([H])C([H])([H])[C@]([H])([C@]([H])(C([H])([H])[H])C([H])([H])C([H])([H])C([H])([H])C([H])(C([H])([H])[H])C([H])([H])[H])[C@@]4(C([H])([H])[H])C([H])([H])C([H])([H])[C@]3([H])[C@@]2(C([H])([H])[H])C([H])([H])C1([H])[H].[H]O[C@]1([H])[C@]([H])(O[H])[C@]([H])(O[C@@]1([H])[N+]1=C([H])C(=C([H])C([H])=C1[H])C(=O)N([H])[H])C([H])([H])OP([O-])(=O)OP([O-])(=O)OC([H])([H])[C@@]1([H])O[C@@]([H])(N2C([H])=NC3=C(N=C([H])N=C23)N([H])[H])[C@]([H])(OP([O-])([O-])=O)[C@]1([H])O[H]
- Finally following command is run:
java -jar rdt-2.4.1-jar-with-dependencies.jar -Q SMI -q '[H]O[C@]1([H])C([H])([H])C2=C([H])C([H])=C3[C@]4([H])C([H])([H])C([H])([H])[C@]([H])([C@]([H])(C([H])([H])[H])C([H])([H])C([H])([H])C([H])([H])C([H])(C([H])([H])[H])C([H])([H])[H])[C@@]4(C([H])([H])[H])C([H])([H])C([H])([H])[C@]3([H])[C@@]2(C([H])([H])[H])C([H])([H])C1([H])[H].[H+].[H]O[C@@]1([H])[C@@]([H])(O[C@]([H])(C([H])([H])OP([O-])(=O)OP([O-])(=O)OC([H])([H])[C@@]2([H])O[C@@]([H])(N3C([H])=NC4=C(N=C([H])N=C34)N([H])[H])[C@]([H])(OP([O-])([O-])=O)[C@]2([H])O[H])[C@@]1([H])O[H])N1C([H])=C([H])C([H])([H])C(=C1[H])C(=O)N([H])[H]>>[H]O[C@]1([H])C([H])([H])C2=C([H])C([H])([H])[C@@]3([H])[C@]4([H])C([H])([H])C([H])([H])[C@]([H])([C@]([H])(C([H])([H])[H])C([H])([H])C([H])([H])C([H])([H])C([H])(C([H])([H])[H])C([H])([H])[H])[C@@]4(C([H])([H])[H])C([H])([H])C([H])([H])[C@]3([H])[C@@]2(C([H])([H])[H])C([H])([H])C1([H])[H].[H]O[C@]1([H])[C@]([H])(O[H])[C@]([H])(O[C@@]1([H])[N+]1=C([H])C(=C([H])C([H])=C1[H])C(=O)N([H])[H])C([H])([H])OP([O-])(=O)OP([O-])(=O)OC([H])([H])[C@@]1([H])O[C@@]([H])(N2C([H])=NC3=C(N=C([H])N=C23)N([H])[H])[C@]([H])(OP([O-])([O-])=O)[C@]1([H])O[H]' -c -g -j AAM -p tests/ -f TEXT
- The output is:
0 [main] INFO net.sf.jnati.deploy.artefact.ConfigManager - Loading global configuration
5 [main] DEBUG net.sf.jnati.deploy.artefact.ConfigManager - Loading defaults: jar:file:/truba/home/sariyuka/reaction-decoder/rdt-2.4.1-jar-with-dependencies.jar!/META-INF/jnati/jnati.default-properties
6 [main] INFO net.sf.jnati.deploy.artefact.ConfigManager - Loading artefact configuration: jniinchi-1.03_1
7 [main] DEBUG net.sf.jnati.deploy.artefact.ConfigManager - Loading instance defaults: jar:file:/truba/home/sariyuka/reaction-decoder/rdt-2.4.1-jar-with-dependencies.jar!/META-INF/jnati/jnati.instance.default-properties
10 [main] INFO net.sf.jnati.deploy.repository.ClasspathRepository - Searching classpath for: jniinchi-1.03_1-LINUX-AMD64
12 [main] INFO net.sf.jnati.deploy.repository.LocalRepository - Searching local repository for: jniinchi-1.03_1-LINUX-AMD64
12 [main] DEBUG net.sf.jnati.deploy.repository.LocalRepository - Artefact path: /truba/home/sariyuka/.jnati/repo/jniinchi/1.03_1/LINUX-AMD64
26 [main] INFO net.sf.jnati.deploy.artefact.ManifestReader - Reading manifest
90 [main] INFO net.sf.jnati.deploy.NativeArtefactLocator - Artefact (jniinchi-1.03_1-LINUX-AMD64) location: /truba/home/sariyuka/.jnati/repo/jniinchi/1.03_1/LINUX-AMD64
90 [main] DEBUG net.sf.jnati.deploy.NativeLibraryLoader - Loading library: /truba/home/sariyuka/.jnati/repo/jniinchi/1.03_1/LINUX-AMD64/libJniInchi-1.03_1-LINUX-AMD64.so
Mapped RXN File /truba/home/sariyuka/reaction-decoder/tests/_ECBLAST_smiles_AAM.rxn
Annotated RXN Image /truba/home/sariyuka/reaction-decoder/tests/_ECBLAST_smiles_AAM.png
Output is presented in text format: /truba/home/sariyuka/reaction-decoder/tests/_ECBLAST_smiles_AAM.txt
In this case, all seems good. But the image of mapping completely different from supported by VMH. The image of mapping:
I ran different versions of RDT and tried different options, to see if it's because of the version differences. However, I could not reach the same output with VMH.
There may be a problem with input SMILES notations. Specifically, I think there might be a problem with hydrogen atoms in SMILES.
For RDT-AAM, should canonical SMILES of metabolites be given? If there is a problem with SMILES that i use, how can I solve the metabolite SMILES problem?
Thanks for help, Best...
In the first case you have a PO3H vs PO3- in the second. It can't use that H if it didn't exist....
Also when you say completely different, the input order of the atoms can determine the orientation of the structures when drawn from SMILES. In that case then yes canonicalising first would help but in this case it looks like there were different inputs so the canonical SMILES would still be different. Also note that mapping numbers mean nothing, it's the relationship between them that matters.
For example:
[CH3:3][CH2:2].[OH:1]>>[CH3:3][CH2:2][OH:1]
[OH:1].[CH3:3][CH2:2]>>[CH3:3][CH2:2][OH:1]
mean the same thing but you may get different colors.
In this case, they may not have given the SMILES that have been used for mapping on their web page or datasets. I see what you mean about mapping numbers. It was very helpful.
Thank you so much!!! 👍
Likely the SMILES is normalised to physiological pH, e.g. -CO2H => -CO2–
I understand. So, do you suggest a tool that can normalize reaction smiles as in physiological conditions or a database that offers normalized rxn files?