dynamo-release
 dynamo-release copied to clipboard
dynamo-release copied to clipboard
Question about rank acceleration
Hi, I have a simple questions about rank acceleration. Is abs just changing the sign? If yes, Why is BIK ahead of S100A6 in column 1 of rank_abs_acceleration as follows:
adata.uns['rank_abs_acceleration']
0 1 2 3 4
0 BIK BIK TUBA1B MALAT1 HSPA5
1 HMGB2 FOS HMGB2 BIK HMGB2
2 TMSB4X S100A6 TUBB TUBA1B MALAT1
3 RBP1 PRDX1 TYMS CD24 PTTG1
4 MEST KLF4 BIRC5 HMGB2 ACTG1
... ... ... ... ... ...
1995 CCDC32 ENSA SIK3 SIK3 GRIN3A
1996 TANC1 POU6F2 RIMS1 CRMP1 HDAC9
1997 INTS6 CCDC32 HDAC9 GRIN3A IQCG
1998 ADGRV1 HDAC9 PDE4D CCDC32 RAI1
1999 HDAC9 SRSF1 CRMP1 HDAC9 TANC1
adata.uns['rank_acceleration']
0 1 2 3 4
0 HMGB2 HMGB2 CD24 MALAT1 MALAT1
1 MALAT1 TOP2A ASCL1 TOP2A NEAT1
2 TOP2A CKS2 BNIP3 C6orf48 MIAT
3 CKS2 DDIT3 RPS18 ZFAS1 CDK1
4 EIF4A2 MALAT1 RPL13A EIF4A2 SYT1
... ... ... ... ... ...
1995 BASP1 CD24 PCNA RBP1 PTMS
1996 TFF3 TMSB4X TYMS PRDX1 UBE2S
1997 MEST RBP1 TUBB PCP4 ACTG1
1998 RBP1 BIK HMGB2 TUBA1B PTTG1
1999 TMSB4X S100A6 TUBA1B BIK HSPA5
Can I get the value used by the rank, as I don't know if it is positive or negative regulation. I found the acceleration stored in adata.layers['acceleration'] (cell by gene). Are the rank acceleration ranked by the average of the same group?
Please also solve the #304, if you are free. Thanks!
Hi @lh12565 I am aware of your issues. Will find sometime later tonight to address them
Thanks again for using dynamo. I finally got some time to address your questions:
- Is abs just changing the sign? If yes, Why is BIK ahead of S100A6 in column 1 of rank_abs_acceleration as follows:
- Can I get the value used by the rank, as I don't know if it is positive or negative regulation. 3.Are the rank acceleration ranked by the average of the same group?
My answers are the following:
- yes, the
absmeans that we will first calculate the absolute value of the acceleration (convert negative values to positive values) before we rank genes. This will reveal the magnitute of RNA acceleration while ignore the direction. For your specific question regarding this two genes. I recommend you to plot the acceleration values of this two genes to reveal the acceleration values across cells:dyn.pl.scatters(adata, layer='acceleration', color=['Tmsb4x', 'Cck']) - yes, you can. Please pass the
output_values = Truetodyn.vf.rank_acceleration_genes - that is right, if you pass the cell
groupsargument, you will get the ranking for each cell group. Otherwise it will be ranked across all cells. all ranking are based on the average values.
Btw, I have updated the documentation of the dynamo in my latest commits, please git pull and setup install to see those updates. I will release a new version of dynamo in a few days to pypi.
Please see my answer to your question in the other github issue
Thanks, when I add output_values = True, I find the value of rank_acceleration or rank_abs_acceleration are different, see below, so the sorting is inconsistent for me. I don't know if that's normal.
adata.uns['rank_abs_acceleration']
0 0_values 1 1_values ... 3 3_values 4 4_values
0 BIK 0.000890 BIK 2.094293e-04 ... MALAT1 0.001347 HSPA5 0.002405
1 HMGB2 0.000843 FOS 2.088793e-04 ... BIK 0.001296 HMGB2 0.001846
2 TMSB4X 0.000821 S100A6 1.846818e-04 ... TUBA1B 0.000940 MALAT1 0.001751
3 RBP1 0.000813 PRDX1 1.764531e-04 ... CD24 0.000717 PTTG1 0.001726
4 MEST 0.000782 KLF4 1.518182e-04 ... HMGB2 0.000694 ACTG1 0.001704
... ... ... ... ... ... ... ... ... ...
1995 CCDC32 0.000004 ENSA 9.541227e-07 ... SIK3 0.000003 GRIN3A 0.000004
1996 TANC1 0.000004 POU6F2 9.290505e-07 ... CRMP1 0.000003 HDAC9 0.000004
1997 INTS6 0.000004 CCDC32 8.875585e-07 ... GRIN3A 0.000003 IQCG 0.000004
1998 ADGRV1 0.000004 HDAC9 8.821968e-07 ... CCDC32 0.000003 RAI1 0.000004
1999 HDAC9 0.000004 SRSF1 8.421325e-07 ... HDAC9 0.000003 TANC1 0.000003
adata.uns['rank_acceleration']
0 0_values 1 1_values ... 3 3_values 4 4_values
0 HMGB2 0.000574 HMGB2 0.000122 ... MALAT1 0.001086 MALAT1 0.001613
1 MALAT1 0.000513 TOP2A 0.000109 ... TOP2A 0.000428 NEAT1 0.000807
2 TOP2A 0.000504 CKS2 0.000099 ... C6orf48 0.000409 MIAT 0.000722
3 CKS2 0.000499 DDIT3 0.000094 ... ZFAS1 0.000383 CDK1 0.000586
4 EIF4A2 0.000482 MALAT1 0.000093 ... EIF4A2 0.000381 SYT1 0.000539
... ... ... ... ... ... ... ... ... ...
1995 BASP1 -0.000604 CD24 -0.000099 ... RBP1 -0.000550 PTMS -0.001502
1996 TFF3 -0.000605 TMSB4X -0.000103 ... PRDX1 -0.000552 UBE2S -0.001530
1997 MEST -0.000729 RBP1 -0.000104 ... PCP4 -0.000619 ACTG1 -0.001648
1998 RBP1 -0.000743 BIK -0.000139 ... TUBA1B -0.000700 PTTG1 -0.001653
1999 TMSB4X -0.000786 S100A6 -0.000151 ... BIK -0.001245 HSPA5 -0.002334
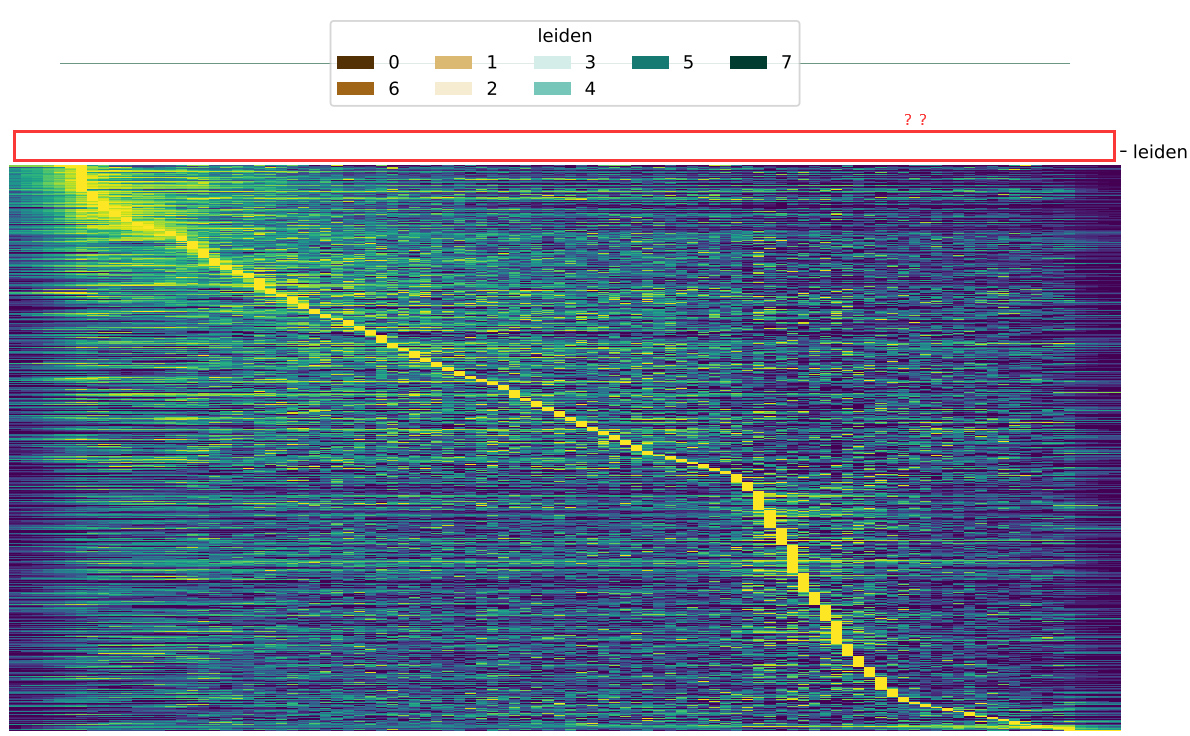
I also have a problem of downstream analysis. When I run the code as follows to visualize the bar of group, the bar did not show up:
dyn.pl.kinetic_heatmap(adata,
genes=transition_genes,
tkey='pca_ddhodge_potential',
gene_order_method='maximum',
mode='pseudotime',
color_map='viridis',
#xticklabels=True,
yticklabels=False,
cell_group='leiden',
cell_group_cmap='BrBG')
Dear @lh12565 , I have explained to you abs means we use the absolute value for ranking. Without abs it of course uses the raw values and that is why you can see negative values in the resultant ranking table.
regarding to the missing leiden group, that is strange because col_colors=col_colors, has passed to the clustermap function which in theory should plot the plot. Have you tried in a different dataset and tried supplied the argument cell_group_cmap with a different color map
@dummyindex could you please also take a look at this issue?
Sure I will take a look at this color bar issue.
This issue is stale because it has been open 90 days with no activity. Remove stale label or comment or this will be closed in 14 days