scispacy doesn't mark end of entities correctly
This bug is based on the example here: https://spacy.io/usage/linguistic-features/
With spacy 2.3.5,
# This is what I recall how I installed
pip install -U pip setuptools wheel
pip install spacy==2.3.5
python -m spacy download en_core_web_sm
nlp = spacy.load("en_core_web_sm")
doc = nlp("San Francisco considers banning sidewalk delivery robots")
ents = [(e.text, e.start_char, e.end_char, e.label_) for e in doc.ents]
print(ents) ### Spacy and Scispacy differ on this
ent_san = [doc[0].text, doc[0].ent_iob_, doc[0].ent_type_]
ent_francisco = [doc[1].text, doc[1].ent_iob_, doc[1].ent_type_]
ent_considers = [doc[2].text, doc[2].ent_iob_, doc[2].ent_type_]
print(ent_san) # ['San', 'B', 'GPE']
print(ent_francisco) # ['Francisco', 'I', 'GPE']
print(ent_considers) # ['considers', 'O', ''] ### Spacy and Scispacy differ on this
generates:
[('San Francisco', 0, 13, 'GPE')]
['San', 'B', 'GPE']
['Francisco', 'I', 'GPE']
['considers', 'O', '']
whereas scispacy 0.3.0
# This is what I recall how I installed
pip install scispacy==0.3.0
pip install https://s3-us-west-2.amazonaws.com/ai2-s2-scispacy/releases/v0.3.0/en_core_sci_sm-0.3.0.tar.gz
nlp = spacy.load('en_core_sci_sm')
doc = nlp("San Francisco considers banning sidewalk delivery robots")
ents = [(e.text, e.start_char, e.end_char, e.label_) for e in doc.ents]
print(ents) ### Spacy and Scispacy differ on this
ent_san = [doc[0].text, doc[0].ent_iob_, doc[0].ent_type_]
ent_francisco = [doc[1].text, doc[1].ent_iob_, doc[1].ent_type_]
ent_considers = [doc[2].text, doc[2].ent_iob_, doc[2].ent_type_]
print(ent_san) # ['San', 'B', 'GPE']
print(ent_francisco) # ['Francisco', 'I', 'GPE']
print(ent_considers) # ['considers', 'O', ''] ### Spacy and Scispacy differ on this
generates stuff for considers which it shouldn't
[('San Francisco', 0, 13, 'ENTITY'), ('considers', 14, 23, 'ENTITY')]
['San', 'B', 'ENTITY']
['Francisco', 'I', 'ENTITY']
['considers', 'B', 'ENTITY']
also... I don't. know if scispacy should be more specific for type of Entity for "San Francisco", but clearly marking considers as the beginning of an ENTITY is wrong.
The entity extractor in the core scispacy models is trained on the medmentions dataset (https://github.com/chanzuckerberg/MedMentions). This is a very different dataset from the one that the core spacy models are trained on, and will broadly recognize things that can be linked to UMLS (https://www.nlm.nih.gov/research/umls/index.html). This means it will identify seemingly generic terms likeconsiders. Depending on your use case, we have more specific NER models available (https://github.com/allenai/scispacy#available-models), or you may want to filter entities based on their UMLS type (type tree is here: https://ai2-s2-scispacy.s3-us-west-2.amazonaws.com/data/umls_semantic_type_tree.tsv)
Depending on your use case, we have more specific NER models available (https://github.com/allenai/scispacy#available-models)
I tried pip install https://s3-us-west-2.amazonaws.com/ai2-s2-scispacy/releases/v0.3.0/en_core_sci_lg-0.3.0.tar.gz
and got worse (but different) results. considers is no longer an entity but other non-entity words became entities
[('San Francisco', 0, 13, 'ENTITY'), ('banning', 24, 31, 'ENTITY'), ('sidewalk', 32, 40, 'ENTITY'), ('delivery', 41, 49, 'ENTITY')]
And for md model:
pip install https://s3-us-west-2.amazonaws.com/ai2-s2-scispacy/releases/v0.3.0/en_core_sci_md-0.3.0.tar.gz
[('San Francisco', 0, 13, 'ENTITY'), ('banning', 24, 31, 'ENTITY'), ('sidewalk', 32, 40, 'ENTITY'), ('delivery', 41, 49, 'ENTITY'), ('robots', 50, 56, 'ENTITY')]
What I was hoping for, was that scispacy would be better at picking out biomedical entities than spacy. I have a mix of biomedical and non-biomedical text, so I was hoping a differential between the two would be useful.
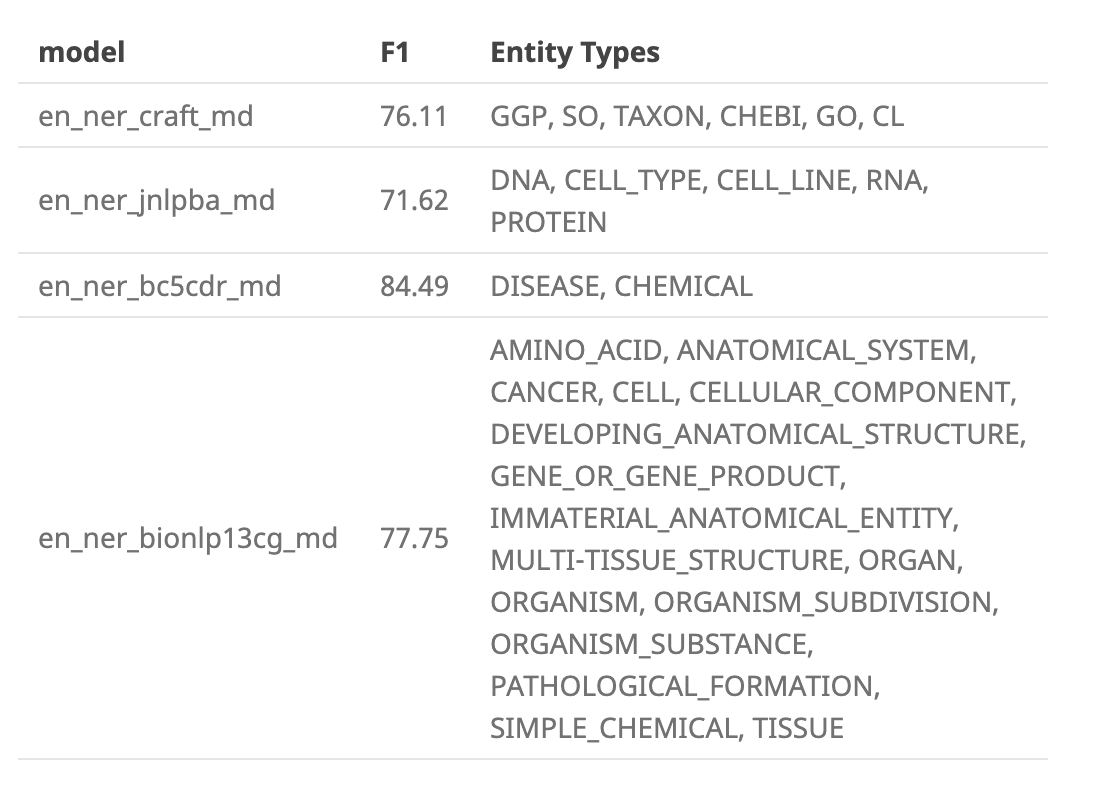
Also... are there vocabulary size estimates for: en_ner_craft_md, en_ner_jnlpba_md, en_ner_bc5cdr_md, en_ner_bionlp13cg_md sizes?
(I'm a bit confused in this transition period between Spacy 2.3.5 -> 3.0.1 and SciSpacy 0.3.0 and 0.4.0. It seems like 2.3.5 goes with 0.3.0 and not 0.4.0).
These are the "more specific NER models" I was referring to. They are each trained on a different biomedical corpus, and have different entity types.

As I said, the core models are trained on the medmentions dataset, which takes a broad definition for what is an entity (and there will be both obvious mistakes, and use case specific mistakes, as this is a trained model). Further filtering can be done using the entity linker and its types.
That being said, the core spacy models are not trained to recognize biomedical entities at all. Here is an example difference
In [27]: sm = spacy.load('en_core_web_sm')
In [28]: sci = spacy.load('en_core_sci_sm')
In [29]: sm_doc = sm("Deoxyribonucleic acid is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organi
...: sms and many viruses.")
In [30]: sci_doc = sci("Deoxyribonucleic acid is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known orga
...: nisms and many viruses.")
In [31]: sm_doc.ents
Out[31]: (two,)
In [32]: sci_doc.ents
Out[32]:
(Deoxyribonucleic acid,
molecule,
polynucleotide chains,
coil,
double helix,
genetic instructions,
development,
functioning,
growth,
reproduction,
organisms,
viruses)
These are the "more specific NER models" I was referring to. They are each trained on a different biomedical corpus, and have different entity types.

As I said, the core models are trained on the medmentions dataset, which takes a broad definition for what is an entity (and there will be both obvious mistakes, and use case specific mistakes, as this is a trained model). Further filtering can be done using the entity linker and its types.
That being said, the core spacy models are not trained to recognize biomedical entities at all. Here is an example difference
In [27]: sm = spacy.load('en_core_web_sm') In [28]: sci = spacy.load('en_core_sci_sm') In [29]: sm_doc = sm("Deoxyribonucleic acid is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organi ...: sms and many viruses.") In [30]: sci_doc = sci("Deoxyribonucleic acid is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known orga ...: nisms and many viruses.") In [31]: sm_doc.ents Out[31]: (two,) In [32]: sci_doc.ents Out[32]: (Deoxyribonucleic acid, molecule, polynucleotide chains, coil, double helix, genetic instructions, development, functioning, growth, reproduction, organisms, viruses)
Hey @danielkingai2 Can I know how to train my own ner extractor on my own corpus from scratch? Basically, I want to train my own "more specific NER models" with my corpus. How do I proceed.. Any reference links that you can share?
we're using spacy 3's new config system and spacy projects. So our project file lives here and our configs live here. You should be able to follow these as a guide, an basically just run the ner training commands, but with your data.
getting this error in pycharm whenever i try to download en_ner_bc5cdr_md ✘ No compatible package found for 'en_ner_bc5cdr_md' (spaCy v3.5.3) python is 3.11