ggtree
 ggtree copied to clipboard
ggtree copied to clipboard
Warning message from `geom_cladelabel`
trafficstars
The fontface parameter gives a warning message when used in geom_cladelabel:
library(ggtree)
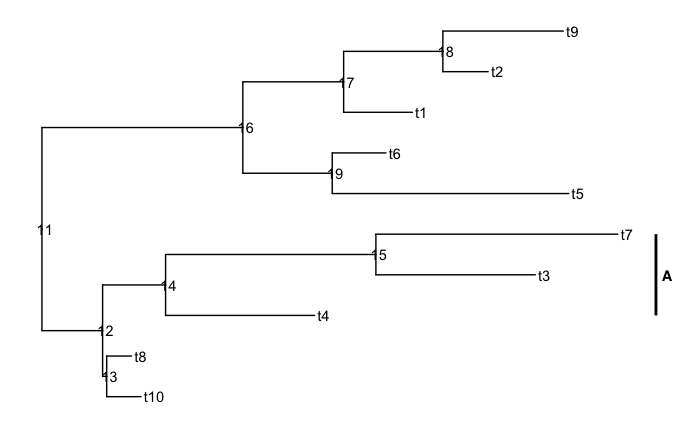
set.seed(3)
ggtree(rtree(10)) +
geom_tiplab() +
geom_nodelab(aes(label = node)) +
geom_cladelabel(node = 14, label = "A", offset = 0.1, barsize = 1, fontface = 2)
Warning: Ignoring unknown parameters: fontface
However, using fontface = 2 does change the label text to bold.
This warning message does not arise when using the fontface parameter in geom_cladelab.
Session information:
R version 4.1.1 (2021-08-10)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Monterey 12.0.1
Matrix products: default
LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ampir_1.1.0 ggtree_3.0.4 randomcoloR_1.1.0.1 broom_0.7.11 ggtext_0.1.1 pals_1.7
[7] patchwork_1.1.1 treeio_1.16.2 ape_5.6-1 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[13] purrr_0.3.4 readr_2.1.1 tidyr_1.1.4 tibble_3.1.6 ggplot2_3.3.5 tidyverse_1.3.1
loaded via a namespace (and not attached):
[1] Rtsne_0.15 colorspace_2.0-2 ellipsis_0.3.2 class_7.3-19 Peptides_2.4.4
[6] fs_1.5.2 aplot_0.1.2 gridtext_0.1.4 dichromat_2.0-0 rstudioapi_0.13
[11] listenv_0.8.0 farver_2.1.0 bit64_4.0.5 prodlim_2019.11.13 fansi_1.0.2
[16] lubridate_1.8.0 xml2_1.3.3 codetools_0.2-18 splines_4.1.1 knitr_1.37
[21] jsonlite_1.7.2 pROC_1.18.0 caret_6.0-90 cluster_2.1.2 dbplyr_2.1.1
[26] BiocManager_1.30.16 mapproj_1.2.8 compiler_4.1.1 httr_1.4.2 backports_1.4.1
[31] assertthat_0.2.1 Matrix_1.3-4 fastmap_1.1.0 lazyeval_0.2.2 cli_3.1.0
[36] htmltools_0.5.2 tools_4.1.1 gtable_0.3.0 glue_1.6.0 reshape2_1.4.4
[41] maps_3.4.0 V8_4.0.0 Rcpp_1.0.8 cellranger_1.1.0 vctrs_0.3.8
[46] nlme_3.1-152 iterators_1.0.13 timeDate_3043.102 xfun_0.29 gower_0.2.2
[51] globals_0.14.0 rvest_1.0.2 lifecycle_1.0.1 future_1.23.0 MASS_7.3-54
[56] scales_1.1.1 ipred_0.9-12 vroom_1.5.7 hms_1.1.1 parallel_4.1.1
[61] yaml_2.2.1 curl_4.3.2 ggfun_0.0.4 yulab.utils_0.0.4 rpart_4.1-15
[66] stringi_1.7.6 foreach_1.5.1 tidytree_0.3.7 lava_1.6.10 rlang_0.4.12
[71] pkgconfig_2.0.3 evaluate_0.14 lattice_0.20-44 recipes_0.1.17 labeling_0.4.2
[76] bit_4.0.4 tidyselect_1.1.1 parallelly_1.30.0 plyr_1.8.6 magrittr_2.0.1
[81] R6_2.5.1 generics_0.1.1 DBI_1.1.2 pillar_1.6.4 haven_2.4.3
[86] withr_2.4.3 survival_3.2-11 nnet_7.3-16 future.apply_1.8.1 modelr_0.1.8
[91] crayon_1.4.2 utf8_1.2.2 tzdb_0.2.0 rmarkdown_2.11 grid_4.1.1
[96] readxl_1.3.1 data.table_1.14.2 ModelMetrics_1.2.2.2 reprex_2.0.1 digest_0.6.29
[101] gridGraphics_0.5-1 stats4_4.1.1 munsell_0.5.0 ggplotify_0.1.0